Near Deterministic Rule Mining and Anomaly Detection in Clinical Data
This notebook contains the complete implementation and usage example of the HierarchicalRulesExplorer class for near-deterministic association rule discovery and anomaly detection in structured data (e.g., clinical data).
The approach combines data discretization, frequent itemset mining (using the Apriori algorithm), and rule filtering to extract highly reliable association rules. These rules can then be used to assess new data for compliance or potential anomalies.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import warnings
import seaborn as sns
# Import tqdm for progress bars.
from tqdm import tqdm
from mlxtend.preprocessing import TransactionEncoder
from mlxtend.frequent_patterns import apriori, association_rules
from pandas.plotting import parallel_coordinates
import networkx as nx
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
436
437
438
439
440
441
442
443
444
445
446
447
448
449
450
451
452
453
454
455
456
457
458
459
460
461
462
463
464
465
466
467
468
469
470
471
472
473
474
475
476
477
478
479
480
481
482
483
484
485
486
487
488
489
490
491
492
493
494
495
496
497
498
499
500
501
502
503
504
505
506
507
508
509
510
511
512
513
514
515
516
517
518
519
520
521
522
523
524
525
526
527
528
529
530
531
532
533
534
535
536
537
538
539
540
541
542
543
544
545
546
547
548
549
550
551
552
553
554
555
556
557
558
559
560
561
562
563
564
565
566
567
568
569
570
571
572
573
574
575
576
577
578
579
580
581
582
583
584
585
586
587
588
589
590
591
592
593
594
595
596
597
598
599
600
601
602
603
604
605
606
607
608
609
610
611
612
613
614
615
616
617
618
619
620
621
622
623
624
625
626
627
628
629
630
631
632
633
634
635
636
637
638
639
640
641
642
643
644
645
646
647
648
649
650
651
652
653
654
655
656
657
658
659
660
661
662
663
664
665
666
667
668
669
670
671
672
673
674
675
676
677
678
679
680
681
682
683
684
685
686
687
688
689
690
691
692
693
694
695
696
697
698
699
700
701
702
703
704
705
706
707
708
709
710
711
712
713
class HierarchicalRulesExplorer:
"""
A class for discovering near-deterministic (almost deterministic) association rules
in a DataFrame and checking the compliance of new data against these rules.
Key features:
- Discretization of continuous variables with custom binning options.
- Extraction and filtering of association rules based on support, confidence, lift, and conviction.
- Methods to summarize, visualize, deduplicate the rules, and evaluate data compliance.
- Enhanced anomaly detection and compliance summary reporting.
- Supports optional sampling and train/validation/test splits.
- Designed for modularity and future GUI integration.
"""
def __init__(self,
min_support=0.01,
min_confidence=0.98,
min_lift=1.0,
min_conviction=0.0,
min_rule_support=None,
deterministic_only=False,
discretization_bins=5,
bin_edges=None,
skip_discretization=None,
min_itemset_length=2,
max_itemset_length=3,
sample_fraction=None, # Optional: fraction of data to sample (0 < sample_fraction <= 1)
display_plots=True, # Flag to display plots (useful for GUI integration)
verbose=True): # Flag to control printing/logging messages
self.min_support = min_support
self.min_confidence = min_confidence
self.min_lift = min_lift
self.min_conviction = min_conviction
self.min_rule_support = min_rule_support if min_rule_support is not None else min_support
self.deterministic_only = deterministic_only
self.discretization_bins = discretization_bins
self.bin_edges = bin_edges if bin_edges is not None else {}
self.skip_discretization = skip_discretization if skip_discretization is not None else []
self.min_itemset_length = min_itemset_length
self.max_itemset_length = max_itemset_length
# New parameters for scalability and interface
self.sample_fraction = sample_fraction
self.display_plots = display_plots
self.verbose = verbose
# Internal storage for discovered rules and binning information
self.rules_ = None
self._trained_bin_info = {}
def _progress(self, iterable, desc="", total=None):
"""
Helper method to wrap an iterable with a progress bar if verbose is True.
Args:
iterable: An iterable to be wrapped.
desc (str): Description for the progress bar.
total (int, optional): Total number of items, if known.
Returns:
iterable: A tqdm-wrapped iterator if verbose is True, else the original iterable.
"""
if self.verbose:
return tqdm(iterable, desc=desc, total=total)
else:
return iterable
def fit(self, df):
"""
Learn near-deterministic association rules from the given DataFrame.
The process includes:
1) (Optional) Sampling the data for scalability.
2) Discretizing continuous columns.
3) Converting each row to a transaction of 'column=value' strings.
4) One-hot encoding and mining frequent itemsets using Apriori.
5) Generating and filtering association rules.
6) Visualizing initial and filtered rule metrics.
7) Reducing redundancy by filtering and deduplicating the rules.
8) (Optionally) Plotting parallel coordinates for the final rule set.
Returns:
self : object
"""
# Optional sampling for large datasets
if self.sample_fraction is not None and 0 < self.sample_fraction < 1:
if self.verbose:
print(f"Sampling data: Using {self.sample_fraction * 100:.1f}% of the input data.")
df = df.sample(frac=self.sample_fraction, random_state=42)
# 1) Discretize continuous columns using training data (fit=True)
df_discretized = self._discretize_continuous_columns(df.copy(), fit=True)
# 2) Convert each row to a transaction of 'column=value' strings.
transactions = self._make_transactions(df_discretized)
# 3) One-hot encode the transactions for frequent pattern mining.
te = TransactionEncoder()
te_array = te.fit(transactions).transform(transactions)
onehot_df = pd.DataFrame(te_array, columns=te.columns_)
# 4) Discover frequent itemsets using Apriori.
itemsets = apriori(
onehot_df,
min_support=self.min_support,
use_colnames=True,
max_len=self.max_itemset_length
)
if itemsets.empty:
warnings.warn("No frequent itemsets found with the specified min_support. "
"Consider lowering the min_support threshold.")
self.rules_ = pd.DataFrame()
return self
# Filter itemsets by the minimum length requirement.
itemsets['length'] = itemsets['itemsets'].apply(len)
itemsets = itemsets[itemsets['length'] >= self.min_itemset_length].reset_index(drop=True)
if itemsets.empty:
warnings.warn("All frequent itemsets are below the min_itemset_length.")
self.rules_ = pd.DataFrame()
return self
# 5) Generate association rules from the frequent itemsets.
rules = association_rules(
itemsets,
metric="confidence",
min_threshold=self.min_confidence
)
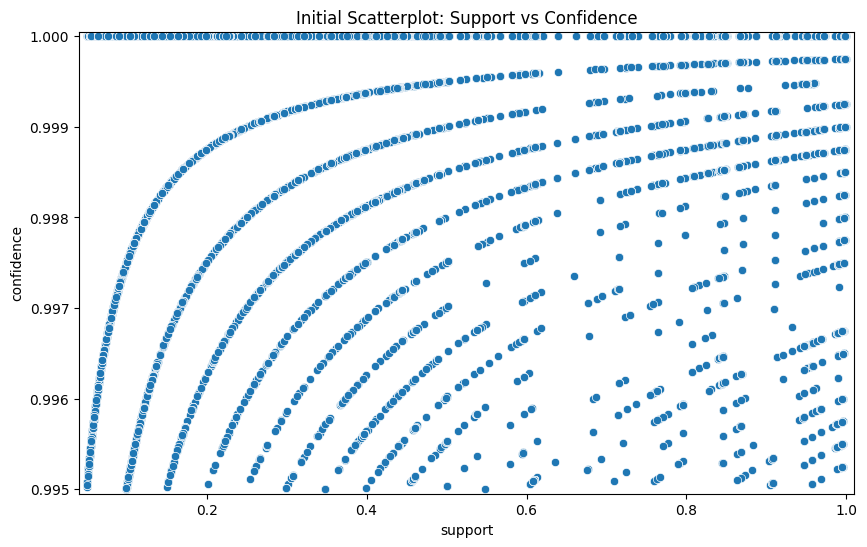
# Initial scatterplot of support vs. confidence (unfiltered rules).
plt.figure(figsize=(10, 6))
sns.scatterplot(x="support", y="confidence", data=rules)
plt.margins(0.01, 0.01)
plt.title("Initial Scatterplot: Support vs Confidence")
if self.display_plots:
plt.show()
else:
plt.close()
# Keep only rules with exactly one item in the consequent.
rules = rules[rules['consequents'].apply(lambda x: len(x) == 1)]
# Filter rules by lift, conviction, and rule support.
rules = rules[(rules['lift'] >= self.min_lift) &
(rules['conviction'] >= self.min_conviction) &
(rules['support'] >= self.min_rule_support)]
# Add a unique rule identifier column for later reference.
rules = rules.reset_index(drop=True)
rules['rule_id'] = rules.index
if rules.empty:
warnings.warn("No association rules were discovered after applying all thresholds. "
"Consider adjusting your parameters.")
self.rules_ = pd.DataFrame()
return self
# Compute an additional 'certainty' column (certainty = 2 * confidence - 1).
rules['certainty'] = 2 * rules['confidence'] - 1
# If only deterministic (certainty==1) rules are desired, filter them.
if self.deterministic_only:
tol = 1e-6 # tolerance for floating point comparisons.
rules = rules[rules['confidence'] >= 1.0 - tol]
if rules.empty:
warnings.warn("No deterministic rules (confidence==1) were found with the current thresholds.")
# Sort rules by confidence and support in descending order.
rules = rules.sort_values(by=['confidence', 'support'], ascending=False).reset_index(drop=True)
self.rules_ = rules
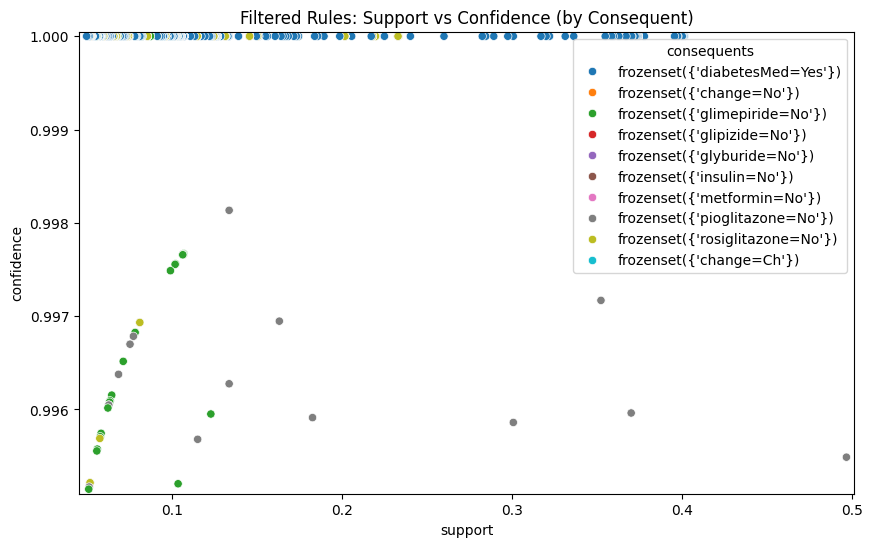
# Scatterplot colored by consequent (filtered rules).
plt.figure(figsize=(10, 6))
sns.scatterplot(x="support", y="confidence", hue="consequents", data=rules)
plt.margins(0.01, 0.01)
plt.title("Filtered Rules: Support vs Confidence (by Consequent)")
if self.display_plots:
plt.show()
else:
plt.close()
# 6) Reduce redundancy: filter maximal rules and deduplicate.
self.filter_maximal_rules()
self.unique_rules()
# 7) Plot parallel coordinates for the final rule set.
#self._plot_parallel_coordinates(self.rules_)
return self
def filter_maximal_rules(self):
"""
For rules sharing the same consequent, keep only those with the largest antecedent set.
This helps remove redundant rules.
"""
if self.rules_ is None or self.rules_.empty:
warnings.warn("No rules available to filter. Ensure the model is fitted first.")
return self.rules_
filtered_groups = []
# Convert groupby iterator to a list to support progress display.
groups = list(self.rules_.groupby('consequents'))
for cons, group in self._progress(groups, desc="Filtering maximal rules", total=len(groups)):
group = group.copy()
indices_to_drop = set()
for i in group.index:
antecedents_i = group.loc[i, 'antecedents']
for j in group.index:
if i == j:
continue
antecedents_j = group.loc[j, 'antecedents']
# If rule i is a proper subset of rule j, mark it for removal.
if antecedents_i.issubset(antecedents_j) and antecedents_i != antecedents_j:
indices_to_drop.add(i)
break
filtered_group = group.drop(index=indices_to_drop)
filtered_groups.append(filtered_group)
self.rules_ = pd.concat(filtered_groups).reset_index(drop=True)
return self.rules_
def unique_rules(self):
"""
Deduplicate rules by ensuring the union of antecedents and consequents is unique.
"""
if self.rules_ is None or self.rules_.empty:
warnings.warn("No rules available to process. Ensure the model is fitted first.")
return self.rules_
self.rules_['rule_itemset'] = self.rules_.apply(
lambda row: frozenset(row['antecedents'].union(row['consequents'])), axis=1
)
self.rules_ = self.rules_.drop_duplicates(subset='rule_itemset')\
.drop(columns=['rule_itemset'])\
.reset_index(drop=True)
return self.rules_
def check_consistency(self, df_test):
"""
Check whether new data violates any of the learned near-deterministic rules.
Returns:
A list of dictionaries, each containing details about a rule violation:
- 'row_index': index of the row in the input DataFrame.
- 'rule_id': identifier of the violated rule.
- 'antecedents': set of antecedents for the rule.
- 'consequents': set of consequents for the rule.
- 'message': description of the violation.
"""
if self.rules_ is None or self.rules_.empty:
raise ValueError("No rules found. Ensure the model is fitted on training data first.")
df_discretized = self._discretize_continuous_columns(df_test.copy(), fit=False)
transactions = self._make_transactions(df_discretized)
inconsistencies = []
for row_index, itemset in enumerate(self._progress(transactions, desc="Checking rule consistency", total=len(transactions))):
itemset_set = set(itemset)
for _, rule in self.rules_.iterrows():
antecedents = set(rule['antecedents'])
consequents = set(rule['consequents'])
if antecedents.issubset(itemset_set) and not consequents.issubset(itemset_set):
inconsistencies.append({
'row_index': row_index,
'rule_id': rule['rule_id'],
'antecedents': antecedents,
'consequents': consequents,
'message': 'Violated near-deterministic rule'
})
return inconsistencies
def check_compliance(self, df_test):
"""
Check compliance of new data against the learned rules and return a new DataFrame with:
- 'compliant': True if no rule is violated; False otherwise.
- 'violated_rules': A list describing the violated rules.
This method is the backbone for both compliance evaluation and anomaly detection.
"""
if self.rules_ is None or self.rules_.empty:
raise ValueError("No rules found. Ensure the model is fitted on training data first.")
df_discretized = self._discretize_continuous_columns(df_test.copy(), fit=False)
transactions = self._make_transactions(df_discretized)
compliance_flags = []
violation_details = []
for row_index, itemset in enumerate(self._progress(transactions, desc="Checking compliance", total=len(transactions))):
itemset_set = set(itemset)
violations = []
for _, rule in self.rules_.iterrows():
antecedents = set(rule['antecedents'])
consequents = set(rule['consequents'])
if antecedents.issubset(itemset_set) and not consequents.issubset(itemset_set):
rule_summary = (f"Rule {rule['rule_id']}: "
f"IF {','.join(sorted(list(antecedents)))} THEN {','.join(sorted(list(consequents)))}")
violations.append(rule_summary)
compliance_flags.append(len(violations) == 0)
violation_details.append(violations)
# Append compliance columns to a copy of the input DataFrame.
df_compliance = df_test.copy()
df_compliance['compliant'] = compliance_flags
df_compliance['violated_rules'] = violation_details
return df_compliance
def evaluate_data(self, df, split_name='Test'):
"""
Evaluate the provided DataFrame against the learned rules.
This method returns both:
- A DataFrame with compliance annotations.
- A summary dictionary with key metrics:
- total_records: Total number of rows evaluated.
- compliant_count: Number of rows compliant with all rules.
- anomaly_count: Number of rows with one or more violations.
- compliance_rate: Proportion of compliant records.
- avg_violations: Average number of violated rules per record.
Args:
df (pd.DataFrame): Data to evaluate.
split_name (str): A label for the data split (e.g., 'Train', 'Validation', 'Test').
Returns:
tuple: (df_compliance, summary_stats)
"""
df_compliance = self.check_compliance(df)
total_records = len(df_compliance)
compliant_count = df_compliance['compliant'].sum()
anomaly_count = total_records - compliant_count
avg_violations = df_compliance['violated_rules'].apply(lambda x: len(x)).mean()
compliance_rate = compliant_count / total_records if total_records > 0 else 0
summary_stats = {
'total_records': total_records,
'compliant_count': compliant_count,
'anomaly_count': anomaly_count,
'compliance_rate': compliance_rate,
'avg_violations': avg_violations
}
if self.verbose:
print(f"\n{split_name} Split Evaluation Summary:")
print(summary_stats)
return df_compliance, summary_stats
def fit_with_validation(self, df_train, df_validation):
"""
Fit the model on the training data and evaluate both training and validation sets.
This method:
1. Fits the rules using the training data.
2. Evaluates compliance on both the training and validation datasets.
Returns:
dict: A dictionary with keys 'train' and 'validation', each mapping to a tuple:
(compliance_dataframe, summary_statistics)
"""
self.fit(df_train)
train_eval = self.evaluate_data(df_train, split_name='Train')
val_eval = self.evaluate_data(df_validation, split_name='Validation')
return {'train': train_eval, 'validation': val_eval}
def detect_anomalies(self, df):
"""
Detect and return only the non-compliant (anomalous) records from the provided DataFrame.
Args:
df (pd.DataFrame): The data to evaluate.
Returns:
pd.DataFrame: Subset of the data that violates one or more rules.
"""
df_compliance, _ = self.evaluate_data(df, split_name='Anomaly Detection')
anomalies = df_compliance[~df_compliance['compliant']]
if self.verbose:
print(f"Detected {len(anomalies)} anomalies out of {len(df_compliance)} records.")
return anomalies
def summarize_rules(self, top_n=10, sort_by='confidence'):
"""
Print a summary of the top association rules.
Args:
top_n (int): Number of top rules to display.
sort_by (str): Column name to sort the rules (default 'confidence').
Returns:
pd.DataFrame: A DataFrame containing the summarized rules.
"""
if self.rules_ is None or self.rules_.empty:
print("No rules to summarize. Fit the model first.")
return None
if sort_by not in self.rules_.columns:
warnings.warn(f"Invalid sort criterion '{sort_by}'. Using 'confidence' by default.")
sort_by = 'confidence'
sorted_rules = self.rules_.sort_values(by=sort_by, ascending=False).head(top_n).copy()
sorted_rules['antecedents'] = sorted_rules['antecedents'].apply(lambda x: ','.join(sorted(list(x))))
sorted_rules['consequents'] = sorted_rules['consequents'].apply(lambda x: ','.join(sorted(list(x))))
cols_to_display = ['rule_id', 'antecedents', 'consequents', 'support', 'confidence', 'lift', 'conviction', 'certainty']
if self.verbose:
print("\nTop Association Rules:")
print(sorted_rules[cols_to_display])
return sorted_rules
def get_rules_summary_statistics(self):
"""
Returns summary statistics (mean, median, std) for rule metrics.
Returns:
pd.DataFrame: Summary statistics for the metrics.
"""
if self.rules_ is None or self.rules_.empty:
print("No rules available to summarize. Fit the model first.")
return pd.DataFrame()
metrics = ['support', 'confidence', 'lift', 'conviction', 'certainty']
stats = {}
for metric in metrics:
stats[metric] = {
'mean': self.rules_[metric].mean(),
'median': self.rules_[metric].median(),
'std': self.rules_[metric].std()
}
stats_df = pd.DataFrame(stats).T
if self.verbose:
print("\nRule Metrics Summary Statistics:")
print(stats_df)
return stats_df
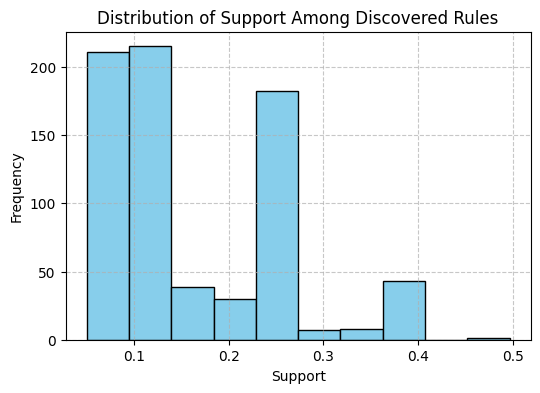
def plot_rule_metric_distribution(self, metric='confidence', bins=10):
"""
Plot the distribution of a specified metric among the discovered rules.
Args:
metric (str): The metric to plot.
bins (int): Number of bins for the histogram.
"""
if self.rules_ is None or self.rules_.empty:
print("No rules to plot. Fit the model first.")
return
if metric not in self.rules_.columns:
print(f"Metric '{metric}' not found in rules. Available columns: {self.rules_.columns.tolist()}")
return
values = self.rules_[metric].dropna()
plt.figure(figsize=(6, 4))
plt.hist(values, bins=bins, edgecolor='k', color='skyblue')
plt.title(f'Distribution of {metric.capitalize()} Among Discovered Rules')
plt.xlabel(metric.capitalize())
plt.ylabel("Frequency")
plt.grid(True, linestyle='--', alpha=0.7)
if self.display_plots:
plt.show()
else:
plt.close()
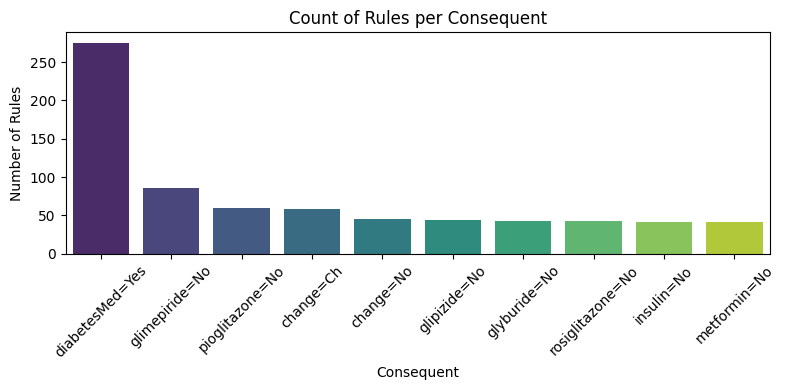
def plot_rules_bar_by_consequent(self):
"""
Plot a bar chart of the count of discovered rules for each consequent.
"""
if self.rules_ is None or self.rules_.empty:
print("No rules available to plot. Fit the model first.")
return
# Create a DataFrame with consequents as string values.
df_rules = self.rules_.copy()
df_rules['consequent_str'] = df_rules['consequents'].apply(lambda x: ','.join(sorted(list(x))))
counts = df_rules['consequent_str'].value_counts()
plt.figure(figsize=(8, 4))
sns.barplot(x=counts.index, y=counts.values, palette="viridis")
plt.title("Count of Rules per Consequent")
plt.xlabel("Consequent")
plt.ylabel("Number of Rules")
plt.xticks(rotation=45)
plt.tight_layout()
if self.display_plots:
plt.show()
else:
plt.close()
def get_rules(self):
"""
Returns the discovered association rules as a DataFrame.
"""
if self.rules_ is not None:
return self.rules_
else:
print("No rules available. Ensure you have called 'fit' first.")
return pd.DataFrame()
# ------------------------ Internal Helper Methods ------------------------
def _make_transactions(self, df_discretized):
"""
Convert each row in the discretized DataFrame to a list of "column=value" strings.
Args:
df_discretized (pd.DataFrame): The discretized data.
Returns:
list: A list of transactions (each transaction is a list of strings).
"""
transactions = []
# Use progress bar for converting rows to transactions.
for _, row in self._progress(df_discretized.iterrows(),
desc="Converting rows to transactions",
total=len(df_discretized)):
items = []
for col in df_discretized.columns:
val = row[col]
if pd.isna(val):
items.append(f"{col}=MISSING")
else:
items.append(f"{col}={val}")
transactions.append(items)
return transactions
def _discretize_continuous_columns(self, df, fit=False):
"""
Discretize numeric columns using custom bin edges or equal-width binning.
Columns listed in skip_discretization are left unchanged.
Args:
df (pd.DataFrame): The input DataFrame.
fit (bool): If True, learn the binning edges; otherwise, use the trained edges.
Returns:
pd.DataFrame: The DataFrame with discretized numeric columns.
"""
numeric_cols = df.select_dtypes(include=[np.number]).columns
# Wrap the loop over numeric columns with a progress bar.
for col in self._progress(numeric_cols, desc="Discretizing numeric columns", total=len(numeric_cols)):
# Convert to object to allow "MISSING" string values.
df[col] = df[col].astype(object)
df.loc[df[col].isna(), col] = "MISSING"
if (col in self.skip_discretization) or all(df[col] == "MISSING"):
continue
if col in self.bin_edges:
edges = self.bin_edges[col]
labels = [f"{col}_bin{i}" for i in range(len(edges) - 1)]
if fit:
self._trained_bin_info[col] = (edges, labels)
else:
edges, labels = self._trained_bin_info[col]
mask = (df[col] != "MISSING")
df.loc[mask, col] = pd.cut(
df.loc[mask, col].astype(float),
bins=edges,
labels=labels,
include_lowest=True
)
else:
if fit:
temp_series = df.loc[df[col] != "MISSING", col].astype(float)
min_val, max_val = temp_series.min(), temp_series.max()
# Handle the case where all values are equal.
if min_val == max_val:
edges = [min_val, min_val + 1e-9]
else:
edges = np.linspace(min_val, max_val, self.discretization_bins + 1)
labels = [f"{col}_bin{i}" for i in range(len(edges) - 1)]
self._trained_bin_info[col] = (edges, labels)
else:
edges, labels = self._trained_bin_info[col]
mask = (df[col] != "MISSING")
df.loc[mask, col] = pd.cut(
df.loc[mask, col].astype(float),
bins=edges,
labels=labels,
include_lowest=True
)
return df
def _plot_parallel_coordinates(self, rules_df):
"""
Private method to plot parallel coordinates for the final set of rules.
Args:
rules_df (pd.DataFrame): DataFrame containing the final set of rules.
"""
if rules_df.empty:
warnings.warn("No rules available for parallel coordinates plot.")
return
# Build a small DataFrame with 'rule_id', 'antecedent', and 'consequent' columns.
pc_data = rules_df[['rule_id', 'antecedents', 'consequents']].copy()
# Convert sets to comma-delimited strings for plotting.
pc_data['antecedent'] = pc_data['antecedents'].apply(lambda x: ','.join(sorted(list(x))))
pc_data['consequent'] = pc_data['consequents'].apply(lambda x: ','.join(sorted(list(x))))
# The parallel_coordinates function requires numeric dimensions; convert categorical labels to codes.
pc_data['antecedent_code'] = pc_data['antecedent'].astype('category').cat.codes
pc_data['consequent_code'] = pc_data['consequent'].astype('category').cat.codes
# Rename 'rule_id' to 'class' for grouping in the plot.
pc_data.rename(columns={'rule_id': 'class'}, inplace=True)
plt.figure(figsize=(5, 8))
parallel_coordinates(pc_data[['class', 'antecedent_code', 'consequent_code']], 'class',
color=sns.color_palette("Spectral", len(pc_data)))
plt.title("Parallel Coordinates: Final Filtered Rules")
plt.grid(True, linestyle='--', alpha=0.7)
plt.xlabel("Features (Antecedent / Consequent Code)")
plt.ylabel("Category Code")
plt.legend([], fontsize=8)
plt.tight_layout()
if self.display_plots:
plt.show()
else:
plt.close()
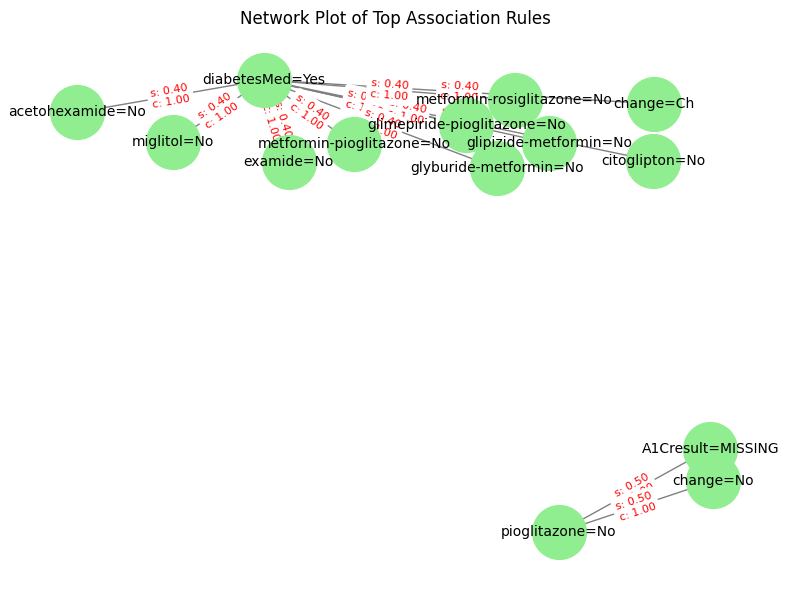
def plot_rules_network(self, top_n=4, support_threshold=0.05, certainty_threshold=0.98):
"""
Plot a network graph of the top association rules based on high support and near-certain confidence.
This method filters the discovered rules to include only those with support above
`support_threshold` and confidence above `certainty_threshold`. It then selects the top_n rules
(sorted by support and confidence) and builds a directed graph where:
- Nodes represent individual items (from either antecedents or consequents).
- Directed edges represent the rule from each antecedent to each consequent.
- Edge labels include the rule's support and confidence.
Args:
top_n (int): Maximum number of rules to display.
support_threshold (float): Minimum support required for a rule to be considered.
certainty_threshold (float): Minimum confidence (e.g., 0.98 for near-certain) required.
Returns:
None
"""
# Ensure that rules have been learned
if self.rules_ is None or self.rules_.empty:
print("No rules available to plot. Ensure the model is fitted first.")
return
# Filter rules based on specified support and confidence thresholds.
filtered_rules = self.rules_[
(self.rules_['support'] >= support_threshold) &
(self.rules_['confidence'] >= certainty_threshold)
]
if filtered_rules.empty:
print("No rules meet the specified support and confidence thresholds.")
return
# Sort rules by support and confidence (in descending order) and select the top_n rules.
filtered_rules = filtered_rules.sort_values(
by=['support', 'confidence'], ascending=False
).head(top_n)
# Initialize a directed graph.
G = nx.DiGraph()
# Iterate over the filtered rules to add nodes and directed edges.
for _, rule in filtered_rules.iterrows():
# Convert antecedents and consequents from sets/frozensets to sorted lists.
antecedents = sorted(rule['antecedents'])
consequents = sorted(rule['consequents'])
# Add nodes for each item.
for item in antecedents:
G.add_node(item, role='antecedent')
for item in consequents:
G.add_node(item, role='consequent')
# Create directed edges from each antecedent to each consequent.
for a in antecedents:
for c in consequents:
# Attach support and confidence as edge attributes.
G.add_edge(a, c, support=rule['support'], confidence=rule['confidence'])
# Use a spring layout for the graph.
pos = nx.spring_layout(G, seed=42) # Seed ensures reproducible layout.
# Plot the graph.
plt.figure(figsize=(8, 6))
nx.draw_networkx_nodes(G, pos, node_color='lightgreen', node_size=1500)
nx.draw_networkx_edges(G, pos, arrowstyle='->', arrowsize=20, edge_color='gray')
nx.draw_networkx_labels(G, pos, font_size=10)
# Create and draw edge labels with support and confidence values.
edge_labels = {
(u, v): f"s: {d['support']:.2f}\nc: {d['confidence']:.2f}"
for u, v, d in G.edges(data=True)
}
nx.draw_networkx_edge_labels(G, pos, edge_labels=edge_labels, font_color='red', font_size=8)
plt.title("Network Plot of Top Association Rules")
plt.axis('off')
plt.tight_layout()
if self.display_plots:
plt.show()
else:
plt.close()
# Attach the new method to the HierarchicalRulesExplorer class.
HierarchicalRulesExplorer.plot_rules_network = plot_rules_network
Extended Discussion
The HierarchicalRulesExplorer class implements a robust pipeline for association rule mining, with a focus on extracting near-deterministic rules that are particularly useful in domains such as clinical data analysis. Here are some key points and motivations behind the design:
- Data Sampling and Preprocessing:
- To handle large datasets, an optional sampling mechanism is available. This ensures that the mining process remains computationally feasible while still capturing the essential patterns.
- Continuous variables are discretized into bins. This step is crucial because many association rule algorithms (such as Apriori) work with categorical data. The option to provide custom bin edges or to use equal-width binning adds flexibility.
- Transaction Encoding:
- Each row in the dataset is converted into a transaction—a list of strings in the form
column=value. This transformation enables the one-hot encoding process that is required for frequent itemset mining.
- Each row in the dataset is converted into a transaction—a list of strings in the form
- Frequent Itemset Mining and Rule Generation:
- The Apriori algorithm is employed to extract frequent itemsets based on a minimum support threshold. This is followed by the generation of association rules that satisfy a high confidence level.
- Additional rule metrics such as lift, conviction, and a computed ‘certainty’ (defined as (2 \times \text{confidence} - 1)) help in filtering out the most reliable rules.
- Rule Filtering and Deduplication:
- To reduce redundancy, the notebook implements a two-step approach:
- Maximal Rule Filtering: For each consequent, only rules with the largest antecedent sets are kept.
- Deduplication: Rules are further deduplicated by ensuring that the union of antecedents and consequents is unique.
- To reduce redundancy, the notebook implements a two-step approach:
- Compliance Checking and Anomaly Detection:
- The learned rules are then used to assess new data. Any record that violates one or more rules is flagged as anomalous. This mechanism can be used for quality control in data streams or as an early-warning system in clinical applications.
This modular design allows for easy integration into larger systems, including potential GUI applications, and offers clear summaries and visualizations at multiple stages of the pipeline.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
# ------------------------------ Example Usage ------------------------------
# Note: You may remove or comment out the following usage example in a production notebook.
if __name__ == "__main__":
# Load the dataset (adjust the file path as needed).
df = pd.read_csv(
'../data/diabetes+130-us+hospitals+for+years+1999-2008/diabetic_data.csv',
na_values='?',
low_memory=True
)
# drop variables that are mostly missing (90 PERCENT)
df = df.dropna(thresh=int(0.1 * len(df)), axis=1)
# Drop columns that are less meaningful for rule discovery.
columns_to_drop = [
'encounter_id',
'patient_nbr',
'admission_type_id',
'discharge_disposition_id',
'admission_source_id'
]
df.drop(columns=columns_to_drop, inplace=True, errors='ignore')
print(df.columns.to_list())
# Split data into training, validation, and testing sets.
df_train = df.iloc[:5000, ].copy()
df_validation = df.iloc[5000:5250, ].copy()
df_test = df.iloc[5250:5500, ].copy()
# Initialize the rules explorer.
explorer = HierarchicalRulesExplorer(
min_support=0.05, # At least 5% of transactions.
min_confidence=0.995, # Nearly deterministic; adjust as needed.
min_lift=1.01, # No extra lift requirement.
min_rule_support=0.02, # Rules must appear in at least 1% of transactions.
deterministic_only=False, # Only show rules with confidence == 1.0.
min_itemset_length=1,
max_itemset_length=3,
sample_fraction=0.8, # Use 80% of the training data (optional)
display_plots=True, # Set to False if integrating into a GUI
verbose=True
)
# 1) Fit the explorer on the training data and evaluate using the validation split.
eval_results = explorer.fit_with_validation(df_train, df_validation)
# 2) Summarize the top discovered (and filtered) rules, sorted by confidence.
explorer.summarize_rules(top_n=10, sort_by='confidence')
# 3) Display summary statistics of the rule metrics.
explorer.get_rules_summary_statistics()
# 4) Plot the distribution of a specified metric among the final rules.
explorer.plot_rule_metric_distribution(metric='support', bins=10)
# 5) Plot a bar chart showing counts of rules per consequent.
explorer.plot_rules_bar_by_consequent()
# 6) Evaluate compliance on a small sample of test data.
df_compliance, test_summary = explorer.evaluate_data(df_test.iloc[:5, :], split_name='Test')
print("\nCompliance DataFrame (first 5 rows):")
print(df_compliance[['compliant', 'violated_rules']])
# 7) Retrieve the rules DataFrame and save to CSV.
rules_df = explorer.get_rules()
print("\nSample of Filtered Rules:\n", rules_df.head(20))
rules_df.to_csv('../data/rules_df.csv', index=False)
# 8) Optionally, detect anomalies (non-compliant records) in the test set.
anomalies = explorer.detect_anomalies(df_test.iloc[:50, :])
print(f"\nDetected {len(anomalies)} anomalies in the test set.")
1
2
3
4
5
6
7
8
9
10
11
12
/var/folders/7z/7gnwr49s6hl4pp9j5dcmgns80000gn/T/ipykernel_56350/127755155.py:6: DtypeWarning: Columns (10) have mixed types. Specify dtype option on import or set low_memory=False.
df = pd.read_csv(
['race', 'gender', 'age', 'time_in_hospital', 'payer_code', 'medical_specialty', 'num_lab_procedures', 'num_procedures', 'num_medications', 'number_outpatient', 'number_emergency', 'number_inpatient', 'diag_1', 'diag_2', 'diag_3', 'number_diagnoses', 'A1Cresult', 'metformin', 'repaglinide', 'nateglinide', 'chlorpropamide', 'glimepiride', 'acetohexamide', 'glipizide', 'glyburide', 'tolbutamide', 'pioglitazone', 'rosiglitazone', 'acarbose', 'miglitol', 'troglitazone', 'tolazamide', 'examide', 'citoglipton', 'insulin', 'glyburide-metformin', 'glipizide-metformin', 'glimepiride-pioglitazone', 'metformin-rosiglitazone', 'metformin-pioglitazone', 'change', 'diabetesMed', 'readmitted']
Sampling data: Using 80.0% of the input data.
Discretizing numeric columns: 100%|██████████| 8/8 [00:00<00:00, 502.23it/s]
Converting rows to transactions: 100%|██████████| 4000/4000 [00:00<00:00, 12851.69it/s]
/Users/denizakdemir/Dropbox/dakdemirGithub/denizakdemir.github.io/.venv/lib/python3.10/site-packages/mlxtend/frequent_patterns/association_rules.py:186: RuntimeWarning: invalid value encountered in divide
cert_metric = np.where(certainty_denom == 0, 0, certainty_num / certainty_denom)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
Filtering maximal rules: 100%|██████████| 10/10 [00:00<00:00, 21.60it/s]
Discretizing numeric columns: 100%|██████████| 8/8 [00:00<00:00, 569.26it/s]
Converting rows to transactions: 100%|██████████| 5000/5000 [00:00<00:00, 12873.46it/s]
Checking compliance: 100%|██████████| 5000/5000 [01:04<00:00, 77.06it/s]
Train Split Evaluation Summary:
{'total_records': 5000, 'compliant_count': np.int64(4961), 'anomaly_count': np.int64(39), 'compliance_rate': np.float64(0.9922), 'avg_violations': np.float64(0.0296)}
Discretizing numeric columns: 100%|██████████| 8/8 [00:00<00:00, 913.49it/s]
Converting rows to transactions: 100%|██████████| 250/250 [00:00<00:00, 11485.33it/s]
Checking compliance: 100%|██████████| 250/250 [00:03<00:00, 78.26it/s]
Validation Split Evaluation Summary:
{'total_records': 250, 'compliant_count': np.int64(248), 'anomaly_count': np.int64(2), 'compliance_rate': np.float64(0.992), 'avg_violations': np.float64(0.008)}
Top Association Rules:
rule_id antecedents \
0 57 acetohexamide=No,change=Ch
491 86 age=[70-80),diabetesMed=No
483 317 diabetesMed=No,gender=Male
484 433 diabetesMed=No,number_diagnoses=number_diagnos...
485 441 diabetesMed=No,readmitted=>30
486 427 diabetesMed=No,num_lab_procedures=num_lab_proc...
487 430 diabetesMed=No,num_medications=num_medications...
488 447 diabetesMed=No,time_in_hospital=time_in_hospit...
489 432 diabetesMed=No,number_diagnoses=number_diagnos...
490 420 diabetesMed=No,medical_specialty=MISSING
consequents support confidence lift conviction certainty
0 diabetesMed=Yes 0.40125 1.0 1.304206 inf 1.0
491 glyburide=No 0.06000 1.0 1.146460 inf 1.0
483 glyburide=No 0.10175 1.0 1.146460 inf 1.0
484 glyburide=No 0.09650 1.0 1.146460 inf 1.0
485 glyburide=No 0.08575 1.0 1.146460 inf 1.0
486 glyburide=No 0.07450 1.0 1.146460 inf 1.0
487 glyburide=No 0.07350 1.0 1.146460 inf 1.0
488 glyburide=No 0.06750 1.0 1.146460 inf 1.0
489 glyburide=No 0.06600 1.0 1.146460 inf 1.0
490 glyburide=No 0.06400 1.0 1.146460 inf 1.0
Rule Metrics Summary Statistics:
mean median std
support 0.155985 0.124750 0.091815
confidence 0.999750 1.000000 0.000907
lift 1.375783 1.304206 0.420797
conviction inf inf NaN
certainty 0.999501 1.000000 0.001813
/Users/denizakdemir/Dropbox/dakdemirGithub/denizakdemir.github.io/.venv/lib/python3.10/site-packages/pandas/core/nanops.py:1016: RuntimeWarning: invalid value encountered in subtract
sqr = _ensure_numeric((avg - values) ** 2)
1
2
3
4
5
/var/folders/7z/7gnwr49s6hl4pp9j5dcmgns80000gn/T/ipykernel_56350/801384969.py:483: FutureWarning:
Passing `palette` without assigning `hue` is deprecated and will be removed in v0.14.0. Assign the `x` variable to `hue` and set `legend=False` for the same effect.
sns.barplot(x=counts.index, y=counts.values, palette="viridis")
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
Discretizing numeric columns: 100%|██████████| 8/8 [00:00<00:00, 1275.93it/s]
Converting rows to transactions: 100%|██████████| 5/5 [00:00<00:00, 9677.67it/s]
Checking compliance: 100%|██████████| 5/5 [00:00<00:00, 70.05it/s]
Test Split Evaluation Summary:
{'total_records': 5, 'compliant_count': np.int64(5), 'anomaly_count': np.int64(0), 'compliance_rate': np.float64(1.0), 'avg_violations': np.float64(0.0)}
Compliance DataFrame (first 5 rows):
compliant violated_rules
5250 True []
5251 True []
5252 True []
5253 True []
5254 True []
Sample of Filtered Rules:
antecedents consequents \
0 (change=Ch, acetohexamide=No) (diabetesMed=Yes)
1 (change=Ch, citoglipton=No) (diabetesMed=Yes)
2 (change=Ch, examide=No) (diabetesMed=Yes)
3 (change=Ch, glimepiride-pioglitazone=No) (diabetesMed=Yes)
4 (change=Ch, glipizide-metformin=No) (diabetesMed=Yes)
5 (change=Ch, glyburide-metformin=No) (diabetesMed=Yes)
6 (change=Ch, metformin-pioglitazone=No) (diabetesMed=Yes)
7 (change=Ch, metformin-rosiglitazone=No) (diabetesMed=Yes)
8 (change=Ch, miglitol=No) (diabetesMed=Yes)
9 (change=Ch, nateglinide=No) (diabetesMed=Yes)
10 (change=Ch, payer_code=MISSING) (diabetesMed=Yes)
11 (change=Ch, chlorpropamide=No) (diabetesMed=Yes)
12 (change=Ch, tolazamide=No) (diabetesMed=Yes)
13 (change=Ch, troglitazone=No) (diabetesMed=Yes)
14 (change=Ch, tolbutamide=No) (diabetesMed=Yes)
15 (change=Ch, number_emergency=number_emergency_... (diabetesMed=Yes)
16 (change=Ch, number_outpatient=number_outpatien... (diabetesMed=Yes)
17 (change=Ch, acarbose=No) (diabetesMed=Yes)
18 (change=Ch, repaglinide=No) (diabetesMed=Yes)
19 (change=Ch, pioglitazone=No) (diabetesMed=Yes)
antecedent support consequent support support confidence lift \
0 0.40125 0.76675 0.40125 1.0 1.304206
1 0.40125 0.76675 0.40125 1.0 1.304206
2 0.40125 0.76675 0.40125 1.0 1.304206
3 0.40125 0.76675 0.40125 1.0 1.304206
4 0.40125 0.76675 0.40125 1.0 1.304206
5 0.40125 0.76675 0.40125 1.0 1.304206
6 0.40125 0.76675 0.40125 1.0 1.304206
7 0.40125 0.76675 0.40125 1.0 1.304206
8 0.40125 0.76675 0.40125 1.0 1.304206
9 0.40125 0.76675 0.40125 1.0 1.304206
10 0.40125 0.76675 0.40125 1.0 1.304206
11 0.40100 0.76675 0.40100 1.0 1.304206
12 0.40100 0.76675 0.40100 1.0 1.304206
13 0.40050 0.76675 0.40050 1.0 1.304206
14 0.40025 0.76675 0.40025 1.0 1.304206
15 0.40000 0.76675 0.40000 1.0 1.304206
16 0.39750 0.76675 0.39750 1.0 1.304206
17 0.39725 0.76675 0.39725 1.0 1.304206
18 0.39550 0.76675 0.39550 1.0 1.304206
19 0.37775 0.76675 0.37775 1.0 1.304206
representativity leverage conviction zhangs_metric jaccard \
0 1.0 0.093592 inf 0.389562 0.523313
1 1.0 0.093592 inf 0.389562 0.523313
2 1.0 0.093592 inf 0.389562 0.523313
3 1.0 0.093592 inf 0.389562 0.523313
4 1.0 0.093592 inf 0.389562 0.523313
5 1.0 0.093592 inf 0.389562 0.523313
6 1.0 0.093592 inf 0.389562 0.523313
7 1.0 0.093592 inf 0.389562 0.523313
8 1.0 0.093592 inf 0.389562 0.523313
9 1.0 0.093592 inf 0.389562 0.523313
10 1.0 0.093592 inf 0.389562 0.523313
11 1.0 0.093533 inf 0.389399 0.522987
12 1.0 0.093533 inf 0.389399 0.522987
13 1.0 0.093417 inf 0.389074 0.522335
14 1.0 0.093358 inf 0.388912 0.522008
15 1.0 0.093300 inf 0.388750 0.521682
16 1.0 0.092717 inf 0.387137 0.518422
17 1.0 0.092659 inf 0.386976 0.518096
18 1.0 0.092250 inf 0.385856 0.515813
19 1.0 0.088110 inf 0.374849 0.492664
certainty kulczynski rule_id
0 1.0 0.761656 57
1 1.0 0.761656 96
2 1.0 0.761656 99
3 1.0 0.761656 102
4 1.0 0.761656 104
5 1.0 0.761656 107
6 1.0 0.761656 119
7 1.0 0.761656 120
8 1.0 0.761656 123
9 1.0 0.761656 124
10 1.0 0.761656 138
11 1.0 0.761493 93
12 1.0 0.761493 149
13 1.0 0.761167 151
14 1.0 0.761004 150
15 1.0 0.760841 135
16 1.0 0.759211 137
17 1.0 0.759048 39
18 1.0 0.757907 144
19 1.0 0.746332 139
Discretizing numeric columns: 100%|██████████| 8/8 [00:00<00:00, 809.20it/s]
Converting rows to transactions: 100%|██████████| 50/50 [00:00<00:00, 12081.06it/s]
Checking compliance: 100%|██████████| 50/50 [00:00<00:00, 75.23it/s]
Anomaly Detection Split Evaluation Summary:
{'total_records': 50, 'compliant_count': np.int64(49), 'anomaly_count': np.int64(1), 'compliance_rate': np.float64(0.98), 'avg_violations': np.float64(0.02)}
Detected 1 anomalies out of 50 records.
Detected 1 anomalies in the test set.
Plot rules network
1
explorer.plot_rules_network(top_n=10, support_threshold=0.2, certainty_threshold=0.990)
Additional Summaries and Future Directions
Summary of Findings
- Rule Discovery:
- The pipeline successfully extracts high-confidence association rules that reflect near-deterministic relationships in the data.
- Metrics such as support, confidence, lift, conviction, and the derived certainty provide multiple lenses for evaluating rule strength.
- Data Compliance and Anomaly Detection:
- The compliance evaluation process enables a quick check on new data, flagging records that violate learned rules.
- This can be directly applied to quality assurance in clinical datasets or monitoring systems where rule deviations may signal critical issues.
- Visual Summaries:
- Scatterplots, histograms, bar charts, and parallel coordinate plots aid in understanding the distribution and relationships among the rules.
Future Directions
- Refinement of Discretization Methods:
- Explore adaptive or data-driven binning techniques to further enhance the quality of discretization for continuous variables.
- Integration with Advanced Visualization Tools:
- Consider incorporating interactive visualization libraries (e.g., Plotly or Bokeh) to allow dynamic exploration of rule metrics.
- Expansion to Other Data Domains:
- While this notebook demonstrates the approach using clinical data, the framework is general and can be extended to other domains such as finance, retail, or IoT sensor data.
This notebook serves as both a demonstration of near-deterministic rule discovery and a starting point for further research and development in anomaly detection systems.