High-Fidelity Synthetic Data Generation
High-Fidelity Synthetic Data Generation: A Comprehensive Framework for Privacy-Preserving Data Synthesis
Introduction
In today’s data-driven world, organizations face a critical challenge: how to leverage data for research, development, and analytics while protecting individual privacy and complying with regulations like GDPR and HIPAA. Synthetic data generation offers a compelling solution by creating artificial datasets that preserve the statistical properties of real data without exposing sensitive information.
This blog post introduces a comprehensive Python framework for generating high-quality synthetic data. Unlike simple random sampling or basic generative models, this framework implements a sophisticated 5-pillar approach that discovers and preserves complex relationships, handles missing data patterns, and ensures statistical fidelity.
Key Features
- 🔍 Automatic Data Analysis: Intelligently classifies columns and discovers relationships
- 🎯 Rule Enforcement: Uses CART models to discover and enforce deterministic rules
- 🕳️ Missingness Modeling: Learns and replicates missing data patterns (MAR/MCAR)
- 🎯 Sequential Generation: Preserves conditional dependencies between columns
- 📊 Quality Assessment: Comprehensive statistical testing and visual reporting
- 🔒 Privacy Protection: Ensures no exact duplicates of original data
The 5-Pillar Approach
Pillar 1: Foundational Data Profiling
The framework automatically classifies each column into three types:
- Continuous: Floating-point numbers (e.g., prices, measurements)
- Integer: Whole numbers representing counts or discrete quantities
- Categorical: Text or numeric values representing distinct classes
Pillar 2: Discovering Deterministic Rules
Using Classification and Regression Trees (CART), the framework identifies columns that can be perfectly predicted from others. For example, it might discover that “customer_type” is determined by “income” levels.
Pillar 3: Analyzing Missingness Patterns
The framework distinguishes between:
- MCAR (Missing Completely At Random): Random missingness
- MAR (Missing At Random): Missingness depends on other variables
Pillar 4: Sequential Generation
Instead of generating all columns independently, the framework uses a topological ordering to generate columns sequentially, preserving conditional dependencies.
Pillar 5: Quality Assurance
Comprehensive evaluation including:
- Statistical tests (KS test for continuous, Chi-squared for categorical)
- Correlation preservation
- Rule compliance verification
- Privacy assessment
Installation and Setup
Step 1: Clone the Repository
First, clone the repository from GitHub:
1
2
# Clone the repository
!git clone https://github.com/dakdemir-nmdp/synth-flow.git
1
fatal: destination path 'synth-flow' already exists and is not an empty directory.
Step 2: Install Dependencies
Install the required Python packages:
1
2
# Install dependencies
!pip install -r synth-flow/requirements.txt
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
Defaulting to user installation because normal site-packages is not writeable
Requirement already satisfied: pandas>=1.5.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from -r synth-flow/requirements.txt (line 1)) (2.2.3)
Requirement already satisfied: numpy>=1.21.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from -r synth-flow/requirements.txt (line 2)) (1.25.2)
Requirement already satisfied: scikit-learn>=1.0.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from -r synth-flow/requirements.txt (line 3)) (1.5.2)
Requirement already satisfied: matplotlib>=3.5.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from -r synth-flow/requirements.txt (line 4)) (3.9.2)
Requirement already satisfied: seaborn>=0.11.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from -r synth-flow/requirements.txt (line 5)) (0.13.2)
Requirement already satisfied: scipy>=1.7.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from -r synth-flow/requirements.txt (line 6)) (1.9.3)
Requirement already satisfied: sdv>=1.0.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from -r synth-flow/requirements.txt (line 7)) (1.24.1)
Requirement already satisfied: ctgan>=0.7.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from -r synth-flow/requirements.txt (line 8)) (0.11.0)
Requirement already satisfied: networkx>=2.8.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from -r synth-flow/requirements.txt (line 9)) (3.4)
Requirement already satisfied: python-dateutil>=2.8.2 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from pandas>=1.5.0->-r synth-flow/requirements.txt (line 1)) (2.9.0.post0)
Requirement already satisfied: pytz>=2020.1 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from pandas>=1.5.0->-r synth-flow/requirements.txt (line 1)) (2024.2)
Requirement already satisfied: tzdata>=2022.7 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from pandas>=1.5.0->-r synth-flow/requirements.txt (line 1)) (2024.2)
Requirement already satisfied: joblib>=1.2.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from scikit-learn>=1.0.0->-r synth-flow/requirements.txt (line 3)) (1.4.2)
Requirement already satisfied: threadpoolctl>=3.1.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from scikit-learn>=1.0.0->-r synth-flow/requirements.txt (line 3)) (3.5.0)
Requirement already satisfied: contourpy>=1.0.1 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from matplotlib>=3.5.0->-r synth-flow/requirements.txt (line 4)) (1.3.0)
Requirement already satisfied: cycler>=0.10 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from matplotlib>=3.5.0->-r synth-flow/requirements.txt (line 4)) (0.12.1)
Requirement already satisfied: fonttools>=4.22.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from matplotlib>=3.5.0->-r synth-flow/requirements.txt (line 4)) (4.54.1)
Requirement already satisfied: kiwisolver>=1.3.1 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from matplotlib>=3.5.0->-r synth-flow/requirements.txt (line 4)) (1.4.7)
Requirement already satisfied: packaging>=20.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from matplotlib>=3.5.0->-r synth-flow/requirements.txt (line 4)) (24.1)
Requirement already satisfied: pillow>=8 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from matplotlib>=3.5.0->-r synth-flow/requirements.txt (line 4)) (10.4.0)
Requirement already satisfied: pyparsing>=2.3.1 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from matplotlib>=3.5.0->-r synth-flow/requirements.txt (line 4)) (3.1.4)
Requirement already satisfied: boto3<2.0.0,>=1.28 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from sdv>=1.0.0->-r synth-flow/requirements.txt (line 7)) (1.35.97)
Requirement already satisfied: botocore<2.0.0,>=1.31 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from sdv>=1.0.0->-r synth-flow/requirements.txt (line 7)) (1.35.97)
Requirement already satisfied: cloudpickle>=2.1.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from sdv>=1.0.0->-r synth-flow/requirements.txt (line 7)) (3.1.0)
Requirement already satisfied: graphviz>=0.13.2 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from sdv>=1.0.0->-r synth-flow/requirements.txt (line 7)) (0.20.3)
Requirement already satisfied: tqdm>=4.29 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from sdv>=1.0.0->-r synth-flow/requirements.txt (line 7)) (4.66.5)
Requirement already satisfied: copulas>=0.12.1 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from sdv>=1.0.0->-r synth-flow/requirements.txt (line 7)) (0.12.3)
Requirement already satisfied: deepecho>=0.7.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from sdv>=1.0.0->-r synth-flow/requirements.txt (line 7)) (0.7.0)
Requirement already satisfied: rdt>=1.17.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from sdv>=1.0.0->-r synth-flow/requirements.txt (line 7)) (1.17.1)
Requirement already satisfied: sdmetrics>=0.21.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from sdv>=1.0.0->-r synth-flow/requirements.txt (line 7)) (0.22.0)
Requirement already satisfied: platformdirs>=4.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from sdv>=1.0.0->-r synth-flow/requirements.txt (line 7)) (4.3.6)
Requirement already satisfied: pyyaml>=6.0.1 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from sdv>=1.0.0->-r synth-flow/requirements.txt (line 7)) (6.0.2)
Requirement already satisfied: torch>=1.13.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from ctgan>=0.7.0->-r synth-flow/requirements.txt (line 8)) (2.4.1)
Requirement already satisfied: jmespath<2.0.0,>=0.7.1 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from boto3<2.0.0,>=1.28->sdv>=1.0.0->-r synth-flow/requirements.txt (line 7)) (1.0.1)
Requirement already satisfied: s3transfer<0.11.0,>=0.10.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from boto3<2.0.0,>=1.28->sdv>=1.0.0->-r synth-flow/requirements.txt (line 7)) (0.10.4)
Requirement already satisfied: urllib3!=2.2.0,<3,>=1.25.4 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from botocore<2.0.0,>=1.31->sdv>=1.0.0->-r synth-flow/requirements.txt (line 7)) (2.2.3)
Requirement already satisfied: plotly>=5.10.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from copulas>=0.12.1->sdv>=1.0.0->-r synth-flow/requirements.txt (line 7)) (5.24.1)
Requirement already satisfied: six>=1.5 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from python-dateutil>=2.8.2->pandas>=1.5.0->-r synth-flow/requirements.txt (line 1)) (1.16.0)
Requirement already satisfied: Faker>=17 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from rdt>=1.17.0->sdv>=1.0.0->-r synth-flow/requirements.txt (line 7)) (37.5.3)
Requirement already satisfied: filelock in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from torch>=1.13.0->ctgan>=0.7.0->-r synth-flow/requirements.txt (line 8)) (3.16.1)
Requirement already satisfied: typing-extensions>=4.8.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from torch>=1.13.0->ctgan>=0.7.0->-r synth-flow/requirements.txt (line 8)) (4.12.2)
Requirement already satisfied: sympy in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from torch>=1.13.0->ctgan>=0.7.0->-r synth-flow/requirements.txt (line 8)) (1.13.3)
Requirement already satisfied: jinja2 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from torch>=1.13.0->ctgan>=0.7.0->-r synth-flow/requirements.txt (line 8)) (3.1.4)
Requirement already satisfied: fsspec in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from torch>=1.13.0->ctgan>=0.7.0->-r synth-flow/requirements.txt (line 8)) (2024.9.0)
Requirement already satisfied: tenacity>=6.2.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from plotly>=5.10.0->copulas>=0.12.1->sdv>=1.0.0->-r synth-flow/requirements.txt (line 7)) (9.0.0)
Requirement already satisfied: MarkupSafe>=2.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from jinja2->torch>=1.13.0->ctgan>=0.7.0->-r synth-flow/requirements.txt (line 8)) (3.0.1)
Requirement already satisfied: mpmath<1.4,>=1.1.0 in /Users/dakdemir/Library/Python/3.10/lib/python/site-packages (from sympy->torch>=1.13.0->ctgan>=0.7.0->-r synth-flow/requirements.txt (line 8)) (1.3.0)
[1m[[0m[34;49mnotice[0m[1;39;49m][0m[39;49m A new release of pip is available: [0m[31;49m24.3.1[0m[39;49m -> [0m[32;49m25.2[0m
[1m[[0m[34;49mnotice[0m[1;39;49m][0m[39;49m To update, run: [0m[32;49mpip3 install --upgrade pip[0m
Step 3: Import Required Libraries
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
import sys
import os
sys.path.append('synth-flow')
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from synthetic_data_generator import SyntheticDataGenerator
import warnings
warnings.filterwarnings('ignore')
# Set random seed for reproducibility
np.random.seed(42)
print("All libraries imported successfully!")
1
All libraries imported successfully!
Example 1: Basic Usage with Sample Data
Let’s start with a simple example using synthetic customer data to demonstrate the basic workflow.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
# Create sample customer data
def create_sample_customer_data(n_samples=1000):
"""Create a sample customer dataset with various relationships"""
np.random.seed(42)
# Generate base features
age = np.random.normal(35, 12, n_samples)
age = np.clip(age, 18, 80).round().astype(int)
# Income is correlated with age
income = 30000 + age * 800 + np.random.normal(0, 10000, n_samples)
income = np.clip(income, 20000, 200000).round().astype(int)
# Education levels
education_levels = ['High School', 'Bachelor', 'Master', 'PhD']
education_probs = [0.3, 0.4, 0.25, 0.05]
education = np.random.choice(education_levels, n_samples, p=education_probs)
# Credit score is determined by age and income (deterministic rule)
credit_score = np.minimum(850, np.maximum(300,
300 + (age - 18) * 5 + (income - 20000) / 1000 + np.random.normal(0, 20, n_samples)
)).round().astype(int)
# Geographic regions
regions = ['North', 'South', 'East', 'West', 'Central']
region = np.random.choice(regions, n_samples)
# Account balance (continuous)
account_balance = np.random.exponential(5000, n_samples)
account_balance = np.round(account_balance, 2)
# Customer type (derived from income - another deterministic rule)
customer_type = np.where(income < 50000, 'Standard',
np.where(income < 100000, 'Premium', 'VIP'))
# Create DataFrame
data = pd.DataFrame({
'age': age,
'income': income,
'education': education,
'credit_score': credit_score,
'region': region,
'account_balance': account_balance,
'customer_type': customer_type
})
# Introduce some missing values with patterns
# Education is more likely to be missing for older customers (MAR)
education_missing_prob = (age - 18) / (80 - 18) * 0.3
education_missing = np.random.random(n_samples) < education_missing_prob
data.loc[education_missing, 'education'] = np.nan
# Account balance is missing completely at random (MCAR)
balance_missing = np.random.random(n_samples) < 0.1
data.loc[balance_missing, 'account_balance'] = np.nan
return data
# Create the sample data
original_data = create_sample_customer_data(1000)
print("Sample customer data created!")
print(f"Shape: {original_data.shape}")
print(f"\nData types:\n{original_data.dtypes}")
print(f"\nMissing values:\n{original_data.isnull().sum()}")
print(f"\nFirst 5 rows:")
original_data.head()
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
Sample customer data created!
Shape: (1000, 7)
Data types:
age int64
income int64
education object
credit_score int64
region object
account_balance float64
customer_type object
dtype: object
Missing values:
age 0
income 0
education 92
credit_score 0
region 0
account_balance 99
customer_type 0
dtype: int64
First 5 rows:
| age | income | education | credit_score | region | account_balance | customer_type | |
|---|---|---|---|---|---|---|---|
| 0 | 41 | 76794 | Bachelor | 483 | North | 6012.11 | Premium |
| 1 | 33 | 65646 | High School | 385 | West | 1833.13 | Premium |
| 2 | 43 | 64996 | Bachelor | 463 | South | 293.95 | Premium |
| 3 | 53 | 65931 | High School | 527 | East | 2100.75 | Premium |
| 4 | 32 | 62582 | Master | 416 | North | 5497.10 | Premium |
Step 1: Initialize and Train the Generator
1
2
3
4
5
6
7
8
9
10
11
# Initialize the generator with custom parameters
generator = SyntheticDataGenerator(
verbose=True, # Print progress messages
cart_max_depth=5, # Maximum depth for CART models
rule_threshold=0.99 # Threshold for deterministic rules
)
# Train the generator on the original data
print("Training the synthetic data generator...")
print("="*70)
generator.fit(original_data)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
Training the synthetic data generator...
======================================================================
Starting synthetic data generation training...
Input data shape: (1000, 7)
Pillar 1: Profiling data and classifying column types...
Column type distribution: {'integer': 3, 'categorical': 3, 'continuous': 1}
Detailed profile: {'age': 'integer', 'income': 'integer', 'education': 'categorical', 'credit_score': 'integer', 'region': 'categorical', 'account_balance': 'continuous', 'customer_type': 'categorical'}
Computing imputation values for future use...
Pillar 2: Discovering deterministic rules with CART models...
Testing rules for column: age
Testing rules for column: income
Testing rules for column: education
Testing rules for column: credit_score
Testing rules for column: region
Testing rules for column: account_balance
Testing rules for column: customer_type
Found deterministic rule! Score: 1.0000
Found 1 deterministic rules
Generation order: ['age', 'income', 'education', 'credit_score', 'region', 'account_balance', 'customer_type']
Pillar 3: Analyzing and modeling missingness patterns...
Analyzing missingness for education (9.2% missing)
Found MAR pattern (max importance: 0.575)
Analyzing missingness for account_balance (9.9% missing)
Found MAR pattern (max importance: 0.517)
Missingness analysis complete: 2 MAR, 0 MCAR patterns
Training completed successfully!
<synthetic_data_generator.SyntheticDataGenerator at 0x337a08ac0>
Step 2: Generate Synthetic Data
1
2
3
4
5
6
7
8
9
10
# Generate synthetic data
n_synthetic_samples = 500
print(f"\nGenerating {n_synthetic_samples} synthetic samples...")
print("="*70)
synthetic_data = generator.generate(n_synthetic_samples)
print(f"\nSynthetic data shape: {synthetic_data.shape}")
print(f"\nFirst 5 synthetic rows:")
synthetic_data.head()
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
Generating 500 synthetic samples...
======================================================================
Generating 500 synthetic samples...
Generating data using sequential modeling approach...
Generating column: age
Generating column: income
Generating column: education
Generating column: credit_score
Generating column: region
Generating column: account_balance
Generating column: customer_type
Post-processing: Enforcing data types and rules...
Step 1: Enforcing data types...
Step 2: Enforcing deterministic rules...
Applied rule for customer_type
Step 3: Applying missingness patterns...
Applied MAR pattern for education (8.4% missing)
Applied MAR pattern for account_balance (11.6% missing)
Generation completed successfully!
Synthetic data shape: (500, 7)
First 5 synthetic rows:
| age | income | education | credit_score | region | account_balance | customer_type | |
|---|---|---|---|---|---|---|---|
| 0 | 45 | 62084 | High School | 473 | South | 1050.64 | Premium |
| 1 | 39 | 60574 | PhD | 463 | South | NaN | Premium |
| 2 | 53 | 82841 | PhD | 541 | Central | NaN | Premium |
| 3 | 20 | 49309 | PhD | 378 | North | 7329.02 | Standard |
| 4 | 22 | 46022 | Bachelor | 343 | South | 775.21 | Standard |
Step 3: Evaluate Quality
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
# Evaluate the quality of synthetic data
print("Evaluating synthetic data quality...")
print("="*70)
evaluation = generator.evaluate()
# Display key metrics
print(f"\n📊 QUALITY METRICS:")
print(f"Overall Quality Score: {evaluation['overall_quality_score']:.3f}")
print(f"Statistical Quality: {evaluation['overall_statistical_quality']:.3f}")
print(f"Privacy Score: {1-evaluation['privacy_assessment']['exact_duplicate_rate']:.3f}")
if 'correlation_preservation' in evaluation:
print(f"Correlation Preservation: {1-evaluation['correlation_preservation']['mean_absolute_difference']:.3f}")
print(f"\n📋 DISCOVERED PATTERNS:")
print(f"Deterministic Rules Found: {len(generator.deterministic_rules)}")
for col, rule in generator.deterministic_rules.items():
print(f" - {col}: {rule['type']} rule with score {rule['score']:.4f}")
print(f"\nMissingness Patterns: {len(generator.missingness_models)}")
for col, model in generator.missingness_models.items():
if model['type'] == 'MAR':
print(f" - {col}: MAR pattern (importance: {model['importance']:.3f})")
else:
print(f" - {col}: MCAR pattern (rate: {model['missing_rate']:.3f})")
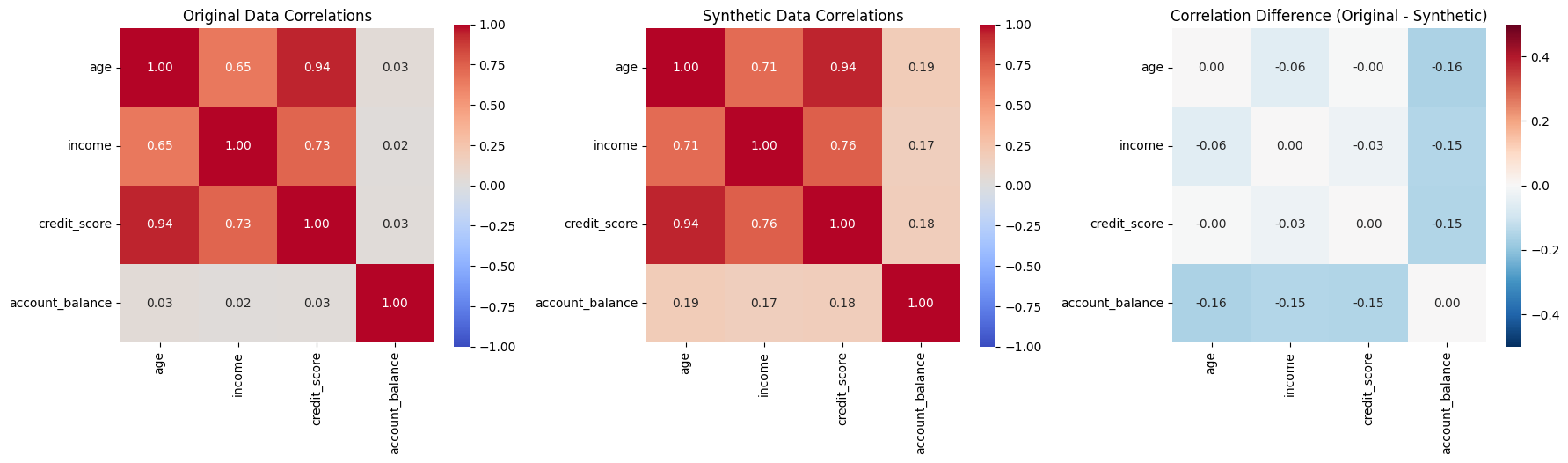
# Display correlation matrices as heatmaps
if 'correlation_preservation' in evaluation:
print("\n📈 CORRELATION ANALYSIS:")
# Extract correlation data
orig_corr_dict = evaluation['correlation_preservation']['original_correlations']
synth_corr_dict = evaluation['correlation_preservation']['synthetic_correlations']
# Get numeric columns
numeric_cols = [col for col, dtype in generator.data_profile.items()
if dtype in ['continuous', 'integer']]
# Convert dictionaries to DataFrames
orig_corr = pd.DataFrame(orig_corr_dict)[numeric_cols].loc[numeric_cols]
synth_corr = pd.DataFrame(synth_corr_dict)[numeric_cols].loc[numeric_cols]
# Create correlation heatmaps
fig, (ax1, ax2, ax3) = plt.subplots(1, 3, figsize=(18, 5))
# Original correlation
sns.heatmap(orig_corr, annot=True, fmt='.2f', cmap='coolwarm', center=0,
square=True, ax=ax1, vmin=-1, vmax=1)
ax1.set_title('Original Data Correlations')
# Synthetic correlation
sns.heatmap(synth_corr, annot=True, fmt='.2f', cmap='coolwarm', center=0,
square=True, ax=ax2, vmin=-1, vmax=1)
ax2.set_title('Synthetic Data Correlations')
# Difference
corr_diff = orig_corr - synth_corr
sns.heatmap(corr_diff, annot=True, fmt='.2f', cmap='RdBu_r', center=0,
square=True, ax=ax3, vmin=-0.5, vmax=0.5)
ax3.set_title('Correlation Difference (Original - Synthetic)')
plt.tight_layout()
plt.show()
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
Evaluating synthetic data quality...
======================================================================
Pillar 5: Evaluating synthetic data quality...
Evaluating statistical fidelity...
Evaluating correlation preservation...
Verifying deterministic rules...
Evaluating missingness fidelity...
Assessing privacy...
Overall Quality Score: 0.976
📊 QUALITY METRICS:
Overall Quality Score: 0.976
Statistical Quality: 0.978
Privacy Score: 1.000
Correlation Preservation: 0.910
📋 DISCOVERED PATTERNS:
Deterministic Rules Found: 1
- customer_type: categorical rule with score 1.0000
Missingness Patterns: 2
- education: MAR pattern (importance: 0.575)
- account_balance: MAR pattern (importance: 0.517)
📈 CORRELATION ANALYSIS:
Step 4: Generate Visual Quality Report
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
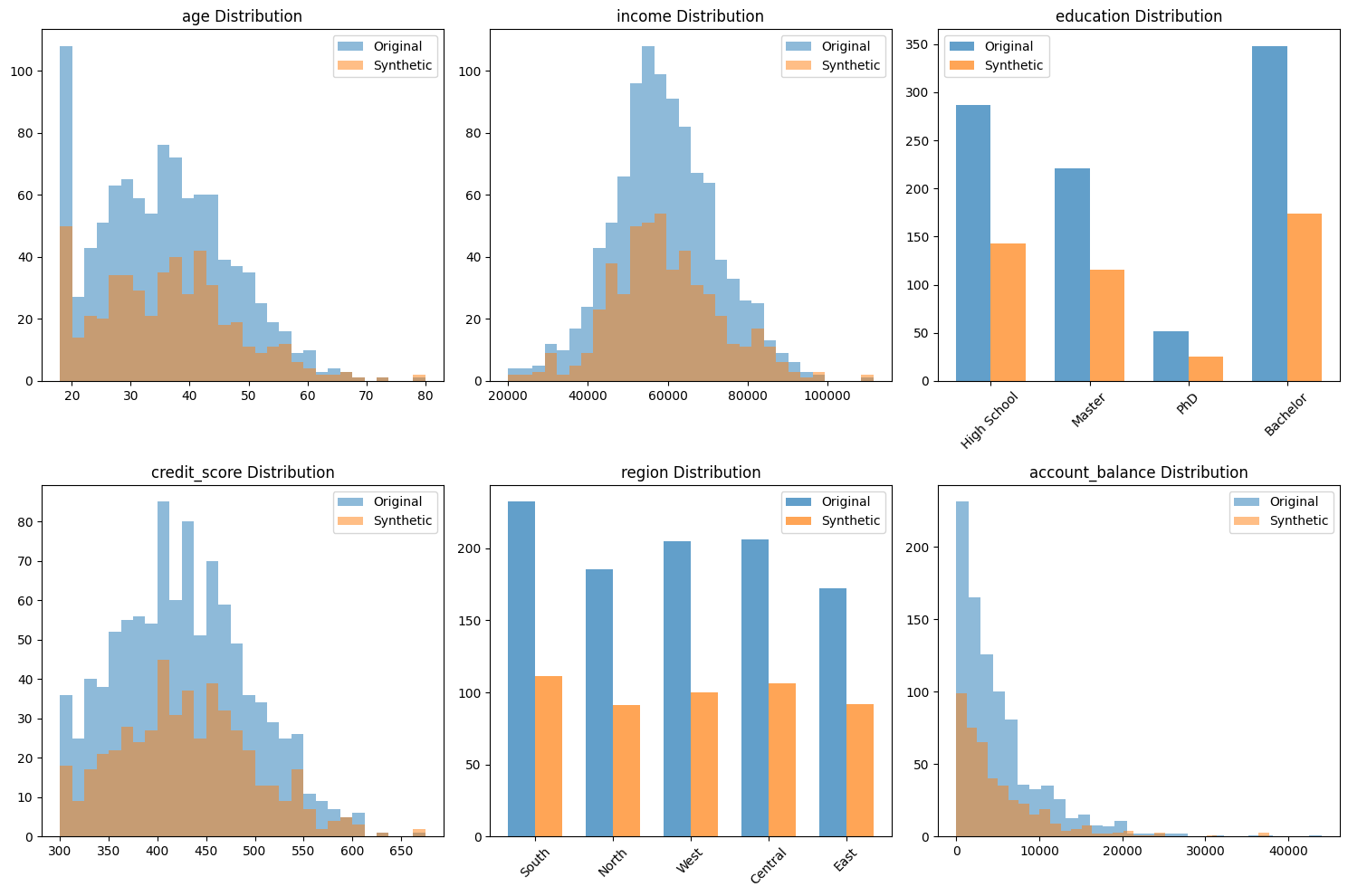
# Generate comprehensive quality report with visualizations
generator.generate_report(save_path="customer_data_quality_report.png")
# Also create a custom comparison visualization
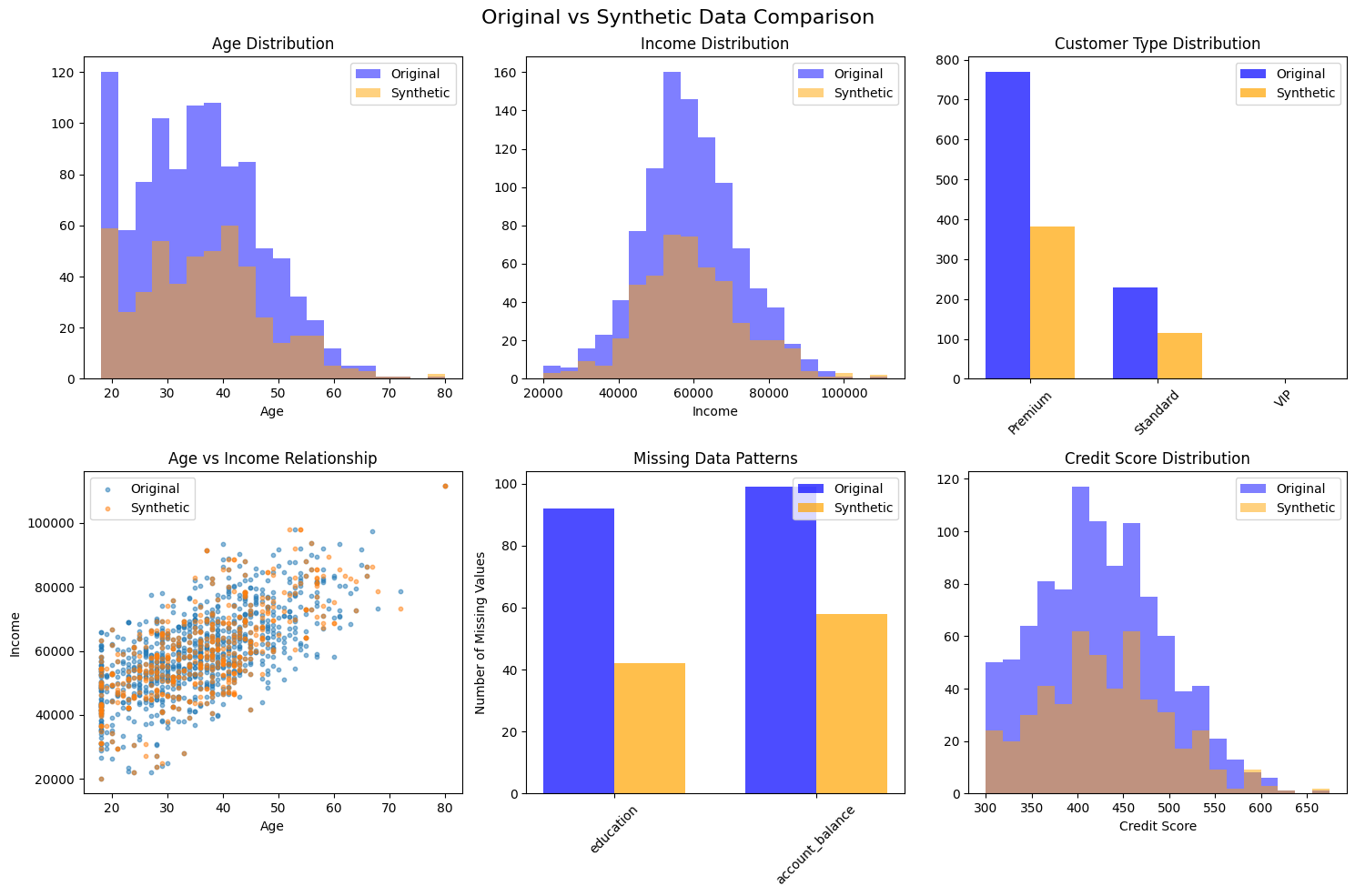
fig, axes = plt.subplots(2, 3, figsize=(15, 10))
fig.suptitle('Original vs Synthetic Data Comparison', fontsize=16)
# 1. Age distribution
axes[0, 0].hist(original_data['age'].dropna(), alpha=0.5, label='Original', bins=20, color='blue')
axes[0, 0].hist(synthetic_data['age'].dropna(), alpha=0.5, label='Synthetic', bins=20, color='orange')
axes[0, 0].set_title('Age Distribution')
axes[0, 0].set_xlabel('Age')
axes[0, 0].legend()
# 2. Income distribution
axes[0, 1].hist(original_data['income'].dropna(), alpha=0.5, label='Original', bins=20, color='blue')
axes[0, 1].hist(synthetic_data['income'].dropna(), alpha=0.5, label='Synthetic', bins=20, color='orange')
axes[0, 1].set_title('Income Distribution')
axes[0, 1].set_xlabel('Income')
axes[0, 1].legend()
# 3. Customer type distribution
orig_counts = original_data['customer_type'].value_counts()
synth_counts = synthetic_data['customer_type'].value_counts()
x = np.arange(len(orig_counts))
width = 0.35
axes[0, 2].bar(x - width/2, orig_counts.values, width, label='Original', alpha=0.7, color='blue')
axes[0, 2].bar(x + width/2, synth_counts.values, width, label='Synthetic', alpha=0.7, color='orange')
axes[0, 2].set_title('Customer Type Distribution')
axes[0, 2].set_xticks(x)
axes[0, 2].set_xticklabels(orig_counts.index, rotation=45)
axes[0, 2].legend()
# 4. Age vs Income scatter
axes[1, 0].scatter(original_data['age'], original_data['income'], alpha=0.5, label='Original', s=10)
axes[1, 0].scatter(synthetic_data['age'], synthetic_data['income'], alpha=0.5, label='Synthetic', s=10)
axes[1, 0].set_title('Age vs Income Relationship')
axes[1, 0].set_xlabel('Age')
axes[1, 0].set_ylabel('Income')
axes[1, 0].legend()
# 5. Missing data comparison
orig_missing = original_data.isnull().sum()
synth_missing = synthetic_data.isnull().sum()
missing_cols = orig_missing[orig_missing > 0].index
if len(missing_cols) > 0:
x_pos = np.arange(len(missing_cols))
axes[1, 1].bar(x_pos - width/2, orig_missing[missing_cols], width, label='Original', alpha=0.7, color='blue')
axes[1, 1].bar(x_pos + width/2, synth_missing[missing_cols], width, label='Synthetic', alpha=0.7, color='orange')
axes[1, 1].set_title('Missing Data Patterns')
axes[1, 1].set_xticks(x_pos)
axes[1, 1].set_xticklabels(missing_cols, rotation=45)
axes[1, 1].set_ylabel('Number of Missing Values')
axes[1, 1].legend()
# 6. Credit score distribution
axes[1, 2].hist(original_data['credit_score'].dropna(), alpha=0.5, label='Original', bins=20, color='blue')
axes[1, 2].hist(synthetic_data['credit_score'].dropna(), alpha=0.5, label='Synthetic', bins=20, color='orange')
axes[1, 2].set_title('Credit Score Distribution')
axes[1, 2].set_xlabel('Credit Score')
axes[1, 2].legend()
plt.tight_layout()
plt.savefig('detailed_comparison.png', dpi=300, bbox_inches='tight')
plt.show()
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
Pillar 5: Evaluating synthetic data quality...
Evaluating statistical fidelity...
Evaluating correlation preservation...
Verifying deterministic rules...
Evaluating missingness fidelity...
Assessing privacy...
Overall Quality Score: 0.976
Quality report saved to: customer_data_quality_report.png
==================================================
SYNTHETIC DATA QUALITY REPORT
==================================================
Overall Quality Score: 0.976
Statistical Quality: 0.978
Correlation Preservation: 0.910
Privacy Score: 1.000
Deterministic Rules: 1 discovered
Rule Compliance: 1.000
Missingness Patterns: 2 analyzed
Column-wise Test Results:
✓ age: KS_test p-value = 0.7169
✓ income: KS_test p-value = 0.9028
✓ education: Chi2_test p-value = 0.9803
✓ credit_score: KS_test p-value = 0.8572
✓ region: Chi2_test p-value = 0.9702
✓ account_balance: KS_test p-value = 0.9735
✓ customer_type: Chi2_test p-value = 0.4685
Example 2: Clinical Data Demonstration
Now let’s demonstrate the framework on real clinical datasets. We’ll use the UCI Hepatitis dataset, which contains various clinical measurements and has natural missing values.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
# Load the Hepatitis dataset
def load_hepatitis_data():
"""Load and prepare the UCI Hepatitis dataset"""
# Define column names based on UCI documentation
column_names = [
'outcome', 'age', 'sex', 'steroid', 'antivirals', 'fatigue', 'malaise',
'anorexia', 'liver_big', 'liver_firm', 'spleen_palpable', 'spiders',
'ascites', 'varices', 'bilirubin', 'alk_phosphate', 'sgot', 'albumin',
'protime', 'histology'
]
# Load the data
data = pd.read_csv('synth-flow/data/hepatitis.csv', names=column_names, na_values='?')
# Convert categorical variables to meaningful labels
binary_map = {1: 'no', 2: 'yes'}
sex_map = {1: 'male', 2: 'female'}
outcome_map = {1: 'die', 2: 'live'}
# Apply mappings
data['outcome'] = data['outcome'].map(outcome_map)
data['sex'] = data['sex'].map(sex_map)
# Binary symptoms/conditions
binary_cols = ['steroid', 'antivirals', 'fatigue', 'malaise', 'anorexia',
'liver_big', 'liver_firm', 'spleen_palpable', 'spiders',
'ascites', 'varices', 'histology']
for col in binary_cols:
data[col] = data[col].map(binary_map)
return data
# Load and analyze the clinical data
try:
clinical_data = load_hepatitis_data()
print("Hepatitis Dataset Loaded Successfully!")
print(f"Shape: {clinical_data.shape}")
print(f"\nMissing values per column:")
missing_summary = clinical_data.isnull().sum()
for col in clinical_data.columns:
if missing_summary[col] > 0:
pct = missing_summary[col] / len(clinical_data) * 100
print(f" {col}: {missing_summary[col]} ({pct:.1f}%)")
print(f"\nOutcome distribution:")
print(clinical_data['outcome'].value_counts())
except FileNotFoundError:
print("Hepatitis dataset not found. Creating simulated clinical data instead...")
# Create simulated clinical data if file not found
n_patients = 155
clinical_data = pd.DataFrame({
'age': np.random.normal(45, 15, n_patients).round().astype(int),
'sex': np.random.choice(['male', 'female'], n_patients, p=[0.7, 0.3]),
'bilirubin': np.random.lognormal(0, 0.5, n_patients),
'albumin': np.random.normal(4.0, 0.5, n_patients),
'outcome': np.random.choice(['live', 'die'], n_patients, p=[0.8, 0.2]),
'fatigue': np.random.choice(['yes', 'no'], n_patients, p=[0.6, 0.4])
})
# Add some missing values
missing_mask = np.random.random((n_patients, 2)) < 0.15
clinical_data.loc[missing_mask[:, 0], 'bilirubin'] = np.nan
clinical_data.loc[missing_mask[:, 1], 'albumin'] = np.nan
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
Hepatitis Dataset Loaded Successfully!
Shape: (155, 20)
Missing values per column:
steroid: 1 (0.6%)
fatigue: 1 (0.6%)
malaise: 1 (0.6%)
anorexia: 1 (0.6%)
liver_big: 10 (6.5%)
liver_firm: 11 (7.1%)
spleen_palpable: 5 (3.2%)
spiders: 5 (3.2%)
ascites: 5 (3.2%)
varices: 5 (3.2%)
bilirubin: 6 (3.9%)
alk_phosphate: 29 (18.7%)
sgot: 4 (2.6%)
albumin: 16 (10.3%)
protime: 67 (43.2%)
Outcome distribution:
outcome
live 123
die 32
Name: count, dtype: int64
Generate Synthetic Clinical Data
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
# Initialize generator for clinical data with adjusted parameters
clinical_generator = SyntheticDataGenerator(
verbose=True,
cart_max_depth=6, # Deeper trees for complex clinical relationships
rule_threshold=0.95 # Slightly relaxed threshold for clinical variability
)
# Train on clinical data
print("\nTraining generator on clinical data...")
print("="*70)
clinical_generator.fit(clinical_data)
# Generate synthetic clinical data
n_synthetic_patients = len(clinical_data)
print(f"\nGenerating {n_synthetic_patients} synthetic patient records...")
synthetic_clinical = clinical_generator.generate(n_synthetic_patients)
print(f"\nSynthetic clinical data generated!")
print(f"First 5 synthetic patient records:")
synthetic_clinical.head()
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
Training generator on clinical data...
======================================================================
Starting synthetic data generation training...
Input data shape: (155, 20)
Pillar 1: Profiling data and classifying column types...
Column type distribution: {'categorical': 14, 'integer': 4, 'continuous': 2}
Detailed profile: {'outcome': 'categorical', 'age': 'integer', 'sex': 'categorical', 'steroid': 'categorical', 'antivirals': 'categorical', 'fatigue': 'categorical', 'malaise': 'categorical', 'anorexia': 'categorical', 'liver_big': 'categorical', 'liver_firm': 'categorical', 'spleen_palpable': 'categorical', 'spiders': 'categorical', 'ascites': 'categorical', 'varices': 'categorical', 'bilirubin': 'continuous', 'alk_phosphate': 'integer', 'sgot': 'integer', 'albumin': 'continuous', 'protime': 'integer', 'histology': 'categorical'}
Computing imputation values for future use...
Pillar 2: Discovering deterministic rules with CART models...
Testing rules for column: outcome
Found deterministic rule! Score: 0.9871
Testing rules for column: age
Testing rules for column: sex
Found deterministic rule! Score: 1.0000
Testing rules for column: steroid
Testing rules for column: antivirals
Found deterministic rule! Score: 0.9746
Testing rules for column: fatigue
Testing rules for column: malaise
Found deterministic rule! Score: 0.9547
Testing rules for column: anorexia
Found deterministic rule! Score: 0.9872
Testing rules for column: liver_big
Found deterministic rule! Score: 0.9932
Testing rules for column: liver_firm
Testing rules for column: spleen_palpable
Found deterministic rule! Score: 0.9589
Testing rules for column: spiders
Testing rules for column: ascites
Found deterministic rule! Score: 1.0000
Testing rules for column: varices
Found deterministic rule! Score: 0.9863
Testing rules for column: bilirubin
Testing rules for column: alk_phosphate
Testing rules for column: sgot
Testing rules for column: albumin
Testing rules for column: protime
Found deterministic rule! Score: 0.9556
Testing rules for column: histology
Warning: Circular dependencies detected, using original column order
Found 10 deterministic rules
Generation order: ['outcome', 'age', 'sex', 'steroid', 'antivirals', 'fatigue', 'malaise', 'anorexia', 'liver_big', 'liver_firm', 'spleen_palpable', 'spiders', 'ascites', 'varices', 'bilirubin', 'alk_phosphate', 'sgot', 'albumin', 'protime', 'histology']
Pillar 3: Analyzing and modeling missingness patterns...
Analyzing missingness for steroid (0.6% missing)
Found MAR pattern (max importance: 0.724)
Analyzing missingness for fatigue (0.6% missing)
Found MAR pattern (max importance: 1.000)
Analyzing missingness for malaise (0.6% missing)
Found MAR pattern (max importance: 1.000)
Analyzing missingness for anorexia (0.6% missing)
Found MAR pattern (max importance: 1.000)
Analyzing missingness for liver_big (6.5% missing)
Found MAR pattern (max importance: 0.903)
Analyzing missingness for liver_firm (7.1% missing)
Found MAR pattern (max importance: 0.903)
Analyzing missingness for spleen_palpable (3.2% missing)
Found MAR pattern (max importance: 0.629)
Analyzing missingness for spiders (3.2% missing)
Found MAR pattern (max importance: 0.629)
Analyzing missingness for ascites (3.2% missing)
Found MAR pattern (max importance: 0.629)
Analyzing missingness for varices (3.2% missing)
Found MAR pattern (max importance: 1.000)
Analyzing missingness for bilirubin (3.9% missing)
Found MAR pattern (max importance: 0.498)
Analyzing missingness for alk_phosphate (18.7% missing)
Found MAR pattern (max importance: 0.564)
Analyzing missingness for sgot (2.6% missing)
Found MAR pattern (max importance: 0.708)
Analyzing missingness for albumin (10.3% missing)
Found MAR pattern (max importance: 0.367)
Analyzing missingness for protime (43.2% missing)
Found MAR pattern (max importance: 0.412)
Missingness analysis complete: 15 MAR, 0 MCAR patterns
Found 1 mixed-type constraints
Training completed successfully!
Generating 155 synthetic patient records...
Generating 155 synthetic samples...
Generating data using sequential modeling approach...
Generating column: outcome
Generating column: age
Generating column: sex
Generating column: steroid
Generating column: antivirals
Generating column: fatigue
Generating column: malaise
Generating column: anorexia
Generating column: liver_big
Generating column: liver_firm
Generating column: spleen_palpable
Generating column: spiders
Generating column: ascites
Generating column: varices
Generating column: bilirubin
Generating column: alk_phosphate
Generating column: sgot
Generating column: albumin
Generating column: protime
Generating column: histology
Post-processing: Enforcing data types and rules...
Step 1: Enforcing data types...
Step 2: Enforcing deterministic rules...
Applied rule for outcome
Applied rule for sex
Applied rule for antivirals
Applied rule for malaise
Applied rule for anorexia
Applied rule for liver_big
Applied rule for spleen_palpable
Applied rule for ascites
Applied rule for varices
Applied rule for protime
Step 3: Applying missingness patterns...
Applied MAR pattern for steroid (0.0% missing)
Applied MAR pattern for fatigue (0.0% missing)
Applied MAR pattern for malaise (0.0% missing)
Applied MAR pattern for anorexia (0.0% missing)
Applied MAR pattern for liver_big (0.0% missing)
Applied MAR pattern for liver_firm (1.9% missing)
Applied MAR pattern for spleen_palpable (0.0% missing)
Applied MAR pattern for spiders (0.0% missing)
Applied MAR pattern for ascites (0.0% missing)
Applied MAR pattern for varices (0.0% missing)
Applied MAR pattern for bilirubin (8.4% missing)
Applied MAR pattern for alk_phosphate (3.2% missing)
Applied MAR pattern for sgot (0.6% missing)
Applied MAR pattern for albumin (7.7% missing)
Applied MAR pattern for protime (37.4% missing)
Step 4: Enforcing mixed-type constraints...
Generation completed successfully!
Synthetic clinical data generated!
First 5 synthetic patient records:
| outcome | age | sex | steroid | antivirals | fatigue | malaise | anorexia | liver_big | liver_firm | spleen_palpable | spiders | ascites | varices | bilirubin | alk_phosphate | sgot | albumin | protime | histology | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | live | 33 | male | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | 0.395644 | 95 | 59 | 4.086249 | 70.0 | no |
| 1 | live | 45 | male | yes | no | yes | yes | yes | yes | yes | yes | yes | yes | yes | 0.519196 | 111 | 145 | NaN | 29.0 | yes |
| 2 | die | 37 | female | yes | no | yes | yes | yes | no | no | yes | yes | yes | yes | 0.400000 | 164 | 53 | 4.146691 | 64.5 | no |
| 3 | live | 37 | male | no | yes | yes | yes | yes | yes | no | yes | yes | yes | yes | 0.233695 | 38 | 90 | 3.317817 | 41.0 | yes |
| 4 | live | 62 | male | no | no | no | yes | yes | yes | no | yes | yes | yes | yes | NaN | 108 | 145 | 3.508119 | 41.0 | yes |
Evaluate Clinical Data Quality
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
# Evaluate synthetic clinical data
clinical_evaluation = clinical_generator.evaluate()
# Create clinical-specific visualizations
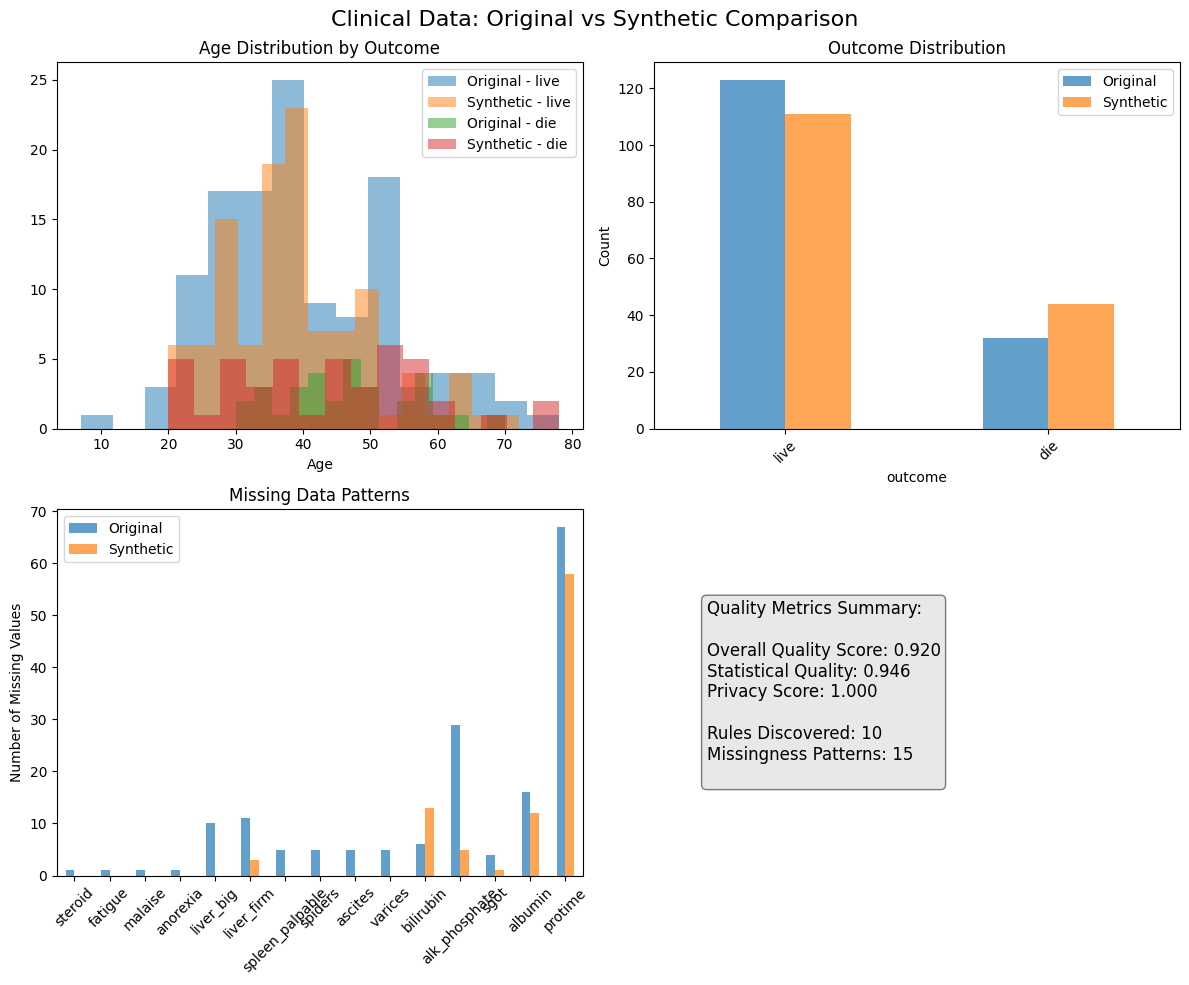
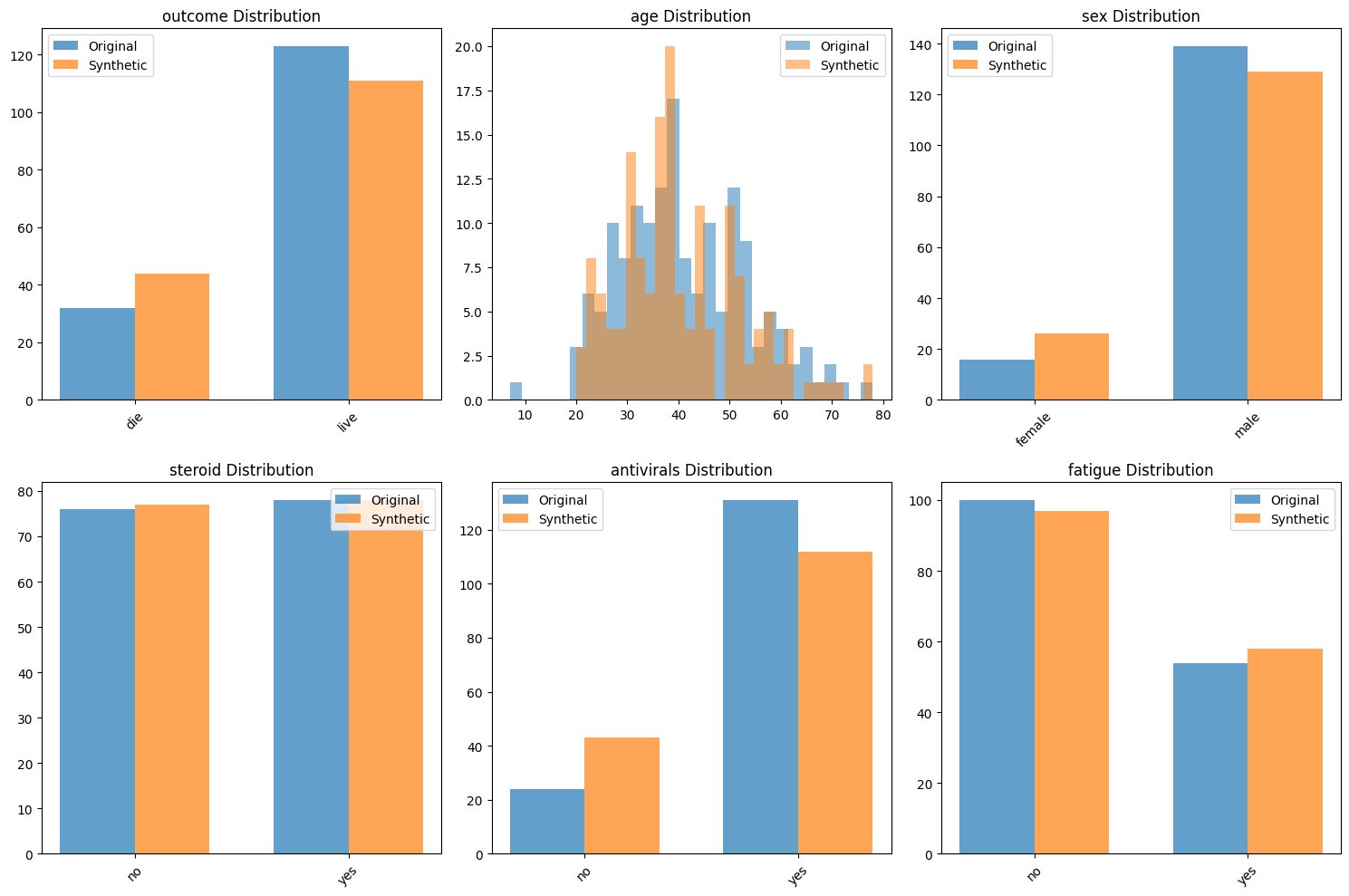
fig, axes = plt.subplots(2, 2, figsize=(12, 10))
fig.suptitle('Clinical Data: Original vs Synthetic Comparison', fontsize=16)
# 1. Age distribution by outcome
for outcome in clinical_data['outcome'].unique():
if pd.notna(outcome):
orig_ages = clinical_data[clinical_data['outcome'] == outcome]['age'].dropna()
axes[0, 0].hist(orig_ages, alpha=0.5, label=f'Original - {outcome}', bins=15)
synth_ages = synthetic_clinical[synthetic_clinical['outcome'] == outcome]['age'].dropna()
axes[0, 0].hist(synth_ages, alpha=0.5, label=f'Synthetic - {outcome}', bins=15, linestyle='--')
axes[0, 0].set_title('Age Distribution by Outcome')
axes[0, 0].set_xlabel('Age')
axes[0, 0].legend()
# 2. Outcome distribution
outcome_comparison = pd.DataFrame({
'Original': clinical_data['outcome'].value_counts(),
'Synthetic': synthetic_clinical['outcome'].value_counts()
})
outcome_comparison.plot(kind='bar', ax=axes[0, 1], alpha=0.7)
axes[0, 1].set_title('Outcome Distribution')
axes[0, 1].set_ylabel('Count')
axes[0, 1].set_xticklabels(axes[0, 1].get_xticklabels(), rotation=45)
# 3. Missing data patterns
missing_comparison = pd.DataFrame({
'Original': clinical_data.isnull().sum(),
'Synthetic': synthetic_clinical.isnull().sum()
})
missing_comparison = missing_comparison[missing_comparison.sum(axis=1) > 0]
if not missing_comparison.empty:
missing_comparison.plot(kind='bar', ax=axes[1, 0], alpha=0.7)
axes[1, 0].set_title('Missing Data Patterns')
axes[1, 0].set_ylabel('Number of Missing Values')
axes[1, 0].set_xticklabels(axes[1, 0].get_xticklabels(), rotation=45)
# 4. Quality metrics summary
axes[1, 1].axis('off')
quality_text = f"""Quality Metrics Summary:
Overall Quality Score: {clinical_evaluation['overall_quality_score']:.3f}
Statistical Quality: {clinical_evaluation['overall_statistical_quality']:.3f}
Privacy Score: {1-clinical_evaluation['privacy_assessment']['exact_duplicate_rate']:.3f}
Rules Discovered: {len(clinical_generator.deterministic_rules)}
Missingness Patterns: {len(clinical_generator.missingness_models)}
"""
axes[1, 1].text(0.1, 0.5, quality_text, fontsize=12, verticalalignment='center',
bbox=dict(boxstyle='round', facecolor='lightgray', alpha=0.5))
plt.tight_layout()
plt.savefig('clinical_data_comparison.png', dpi=300, bbox_inches='tight')
plt.show()
# Display correlation matrices as heatmaps if available
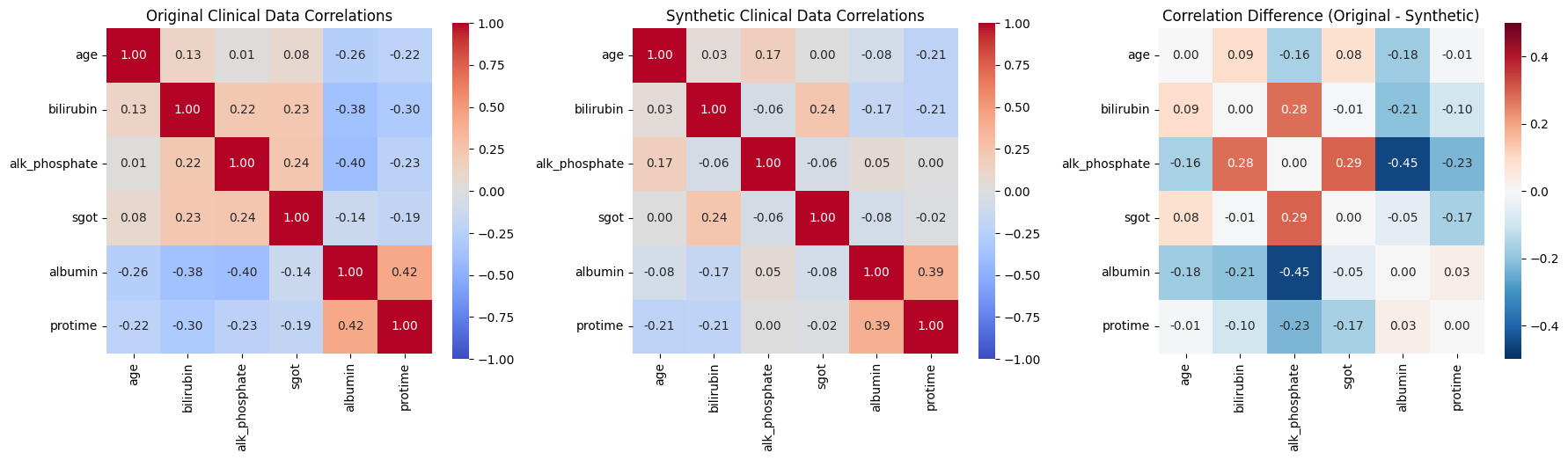
if 'correlation_preservation' in clinical_evaluation:
print("\n📈 CLINICAL DATA CORRELATION ANALYSIS:")
# Extract correlation data
orig_corr_dict = clinical_evaluation['correlation_preservation']['original_correlations']
synth_corr_dict = clinical_evaluation['correlation_preservation']['synthetic_correlations']
# Get numeric columns
numeric_cols = [col for col, dtype in clinical_generator.data_profile.items()
if dtype in ['continuous', 'integer']]
if len(numeric_cols) >= 2:
# Convert dictionaries to DataFrames
orig_corr = pd.DataFrame(orig_corr_dict)[numeric_cols].loc[numeric_cols]
synth_corr = pd.DataFrame(synth_corr_dict)[numeric_cols].loc[numeric_cols]
# Create correlation heatmaps
fig, (ax1, ax2, ax3) = plt.subplots(1, 3, figsize=(18, 5))
# Original correlation
sns.heatmap(orig_corr, annot=True, fmt='.2f', cmap='coolwarm', center=0,
square=True, ax=ax1, vmin=-1, vmax=1)
ax1.set_title('Original Clinical Data Correlations')
# Synthetic correlation
sns.heatmap(synth_corr, annot=True, fmt='.2f', cmap='coolwarm', center=0,
square=True, ax=ax2, vmin=-1, vmax=1)
ax2.set_title('Synthetic Clinical Data Correlations')
# Difference
corr_diff = orig_corr - synth_corr
sns.heatmap(corr_diff, annot=True, fmt='.2f', cmap='RdBu_r', center=0,
square=True, ax=ax3, vmin=-0.5, vmax=0.5)
ax3.set_title('Correlation Difference (Original - Synthetic)')
plt.tight_layout()
plt.show()
# Generate detailed quality report
clinical_generator.generate_report(save_path="clinical_quality_report.png")
1
2
3
4
5
6
7
Pillar 5: Evaluating synthetic data quality...
Evaluating statistical fidelity...
Evaluating correlation preservation...
Verifying deterministic rules...
Evaluating missingness fidelity...
Assessing privacy...
Overall Quality Score: 0.920
1
📈 CLINICAL DATA CORRELATION ANALYSIS:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
Pillar 5: Evaluating synthetic data quality...
Evaluating statistical fidelity...
Evaluating correlation preservation...
Verifying deterministic rules...
Evaluating missingness fidelity...
Assessing privacy...
Overall Quality Score: 0.920
Quality report saved to: clinical_quality_report.png
==================================================
SYNTHETIC DATA QUALITY REPORT
==================================================
Overall Quality Score: 0.920
Statistical Quality: 0.946
Correlation Preservation: 0.843
Privacy Score: 1.000
Deterministic Rules: 10 discovered
Rule Compliance: 0.917
Missingness Patterns: 15 analyzed
Column-wise Test Results:
✓ outcome: Chi2_test p-value = 0.1464
✓ age: KS_test p-value = 0.8316
✓ sex: Chi2_test p-value = 0.1353
✓ steroid: Chi2_test p-value = 1.0000
✗ antivirals: Chi2_test p-value = 0.0130
✓ fatigue: Chi2_test p-value = 0.7549
✓ malaise: Chi2_test p-value = 0.2716
✓ anorexia: Chi2_test p-value = 0.1554
✓ liver_big: Chi2_test p-value = 0.3806
✓ liver_firm: Chi2_test p-value = 0.9417
✓ spleen_palpable: Chi2_test p-value = 0.2849
✓ spiders: Chi2_test p-value = 0.7475
✓ ascites: Chi2_test p-value = 0.5983
✓ varices: Chi2_test p-value = 0.6395
✗ bilirubin: KS_test p-value = 0.0011
✗ alk_phosphate: KS_test p-value = 0.0155
✓ sgot: KS_test p-value = 0.0569
✓ albumin: KS_test p-value = 0.3702
✓ protime: KS_test p-value = 0.0708
✓ histology: Chi2_test p-value = 0.5694
{'column_tests': {'outcome': {'test': 'Chi2_test',
'statistic': 2.1091992802519117,
'p_value': 0.14641591196125164,
'similar': True},
'age': {'test': 'KS_test',
'statistic': 0.07096774193548387,
'p_value': 0.8315953283764764,
'similar': True},
'sex': {'test': 'Chi2_test',
'statistic': 2.2308102345415777,
'p_value': 0.1352828894850494,
'similar': True},
'steroid': {'test': 'Chi2_test',
'statistic': 0.0,
'p_value': 1.0,
'similar': True},
'antivirals': {'test': 'Chi2_test',
'statistic': 6.169154228855722,
'p_value': 0.012999674272798698,
'similar': False},
'fatigue': {'test': 'Chi2_test',
'statistic': 0.0974264858060018,
'p_value': 0.7549400858586965,
'similar': True},
'malaise': {'test': 'Chi2_test',
'statistic': 1.2088833870044255,
'p_value': 0.271553377036263,
'similar': True},
'anorexia': {'test': 'Chi2_test',
'statistic': 2.0180747024367447,
'p_value': 0.15543611485514533,
'similar': True},
'liver_big': {'test': 'Chi2_test',
'statistic': 0.7688642333445797,
'p_value': 0.38056864030085324,
'similar': True},
'liver_firm': {'test': 'Chi2_test',
'statistic': 0.005354946821244084,
'p_value': 0.9416648690620364,
'similar': True},
'spleen_palpable': {'test': 'Chi2_test',
'statistic': 1.1435680785697944,
'p_value': 0.28489963512276645,
'similar': True},
'spiders': {'test': 'Chi2_test',
'statistic': 0.10367040388145825,
'p_value': 0.7474686889957438,
'similar': True},
'ascites': {'test': 'Chi2_test',
'statistic': 0.2775090754526236,
'p_value': 0.5983385240357398,
'similar': True},
'varices': {'test': 'Chi2_test',
'statistic': 0.2194509432560906,
'p_value': 0.6394585908485868,
'similar': True},
'bilirubin': {'test': 'KS_test',
'statistic': 0.22374515549673882,
'p_value': 0.001084109726539591,
'similar': False},
'alk_phosphate': {'test': 'KS_test',
'statistic': 0.18507936507936507,
'p_value': 0.015534185168877704,
'similar': False},
'sgot': {'test': 'KS_test',
'statistic': 0.14978068289326568,
'p_value': 0.05692671393407061,
'similar': True},
'albumin': {'test': 'KS_test',
'statistic': 0.10600191175730744,
'p_value': 0.37020579770920514,
'similar': True},
'protime': {'test': 'KS_test',
'statistic': 0.18544985941893158,
'p_value': 0.07081174939544421,
'similar': True},
'histology': {'test': 'Chi2_test',
'statistic': 0.32367190110257266,
'p_value': 0.5694092780259552,
'similar': True}},
'overall_statistical_quality': 0.9458741296656511,
'correlation_preservation': {'mean_absolute_difference': 0.15652221200114286,
'original_correlations': {'age': {'age': 1.0,
'bilirubin': 0.1254626946933773,
'alk_phosphate': 0.008923785562059456,
'sgot': 0.08254466652180756,
'albumin': -0.26298050964527275,
'protime': -0.22223252154053216},
'bilirubin': {'age': 0.1254626946933773,
'bilirubin': 1.0,
'alk_phosphate': 0.22006186153908958,
'sgot': 0.23296744377722084,
'albumin': -0.37731757035951774,
'protime': -0.3026083058653626},
'alk_phosphate': {'age': 0.008923785562059456,
'bilirubin': 0.22006186153908958,
'alk_phosphate': 1.0,
'sgot': 0.2395813949251737,
'albumin': -0.40385699222122784,
'protime': -0.22684642591560608},
'sgot': {'age': 0.08254466652180756,
'bilirubin': 0.23296744377722084,
'alk_phosphate': 0.2395813949251737,
'sgot': 1.0,
'albumin': -0.1362757629284122,
'protime': -0.18914922175968923},
'albumin': {'age': -0.26298050964527275,
'bilirubin': -0.37731757035951774,
'alk_phosphate': -0.40385699222122784,
'sgot': -0.1362757629284122,
'albumin': 1.0,
'protime': 0.42441920160752206},
'protime': {'age': -0.22223252154053216,
'bilirubin': -0.3026083058653626,
'alk_phosphate': -0.22684642591560608,
'sgot': -0.18914922175968923,
'albumin': 0.42441920160752206,
'protime': 1.0}},
'synthetic_correlations': {'age': {'age': 1.0,
'bilirubin': 0.03326468479946039,
'alk_phosphate': 0.1704030136189319,
'sgot': 0.0007974932531030433,
'albumin': -0.0821222593960426,
'protime': -0.2135133507321673},
'bilirubin': {'age': 0.03326468479946039,
'bilirubin': 1.0,
'alk_phosphate': -0.059951272922023076,
'sgot': 0.24038403856821702,
'albumin': -0.17056491214839042,
'protime': -0.2069968293170332},
'alk_phosphate': {'age': 0.1704030136189319,
'bilirubin': -0.059951272922023076,
'alk_phosphate': 1.0,
'sgot': -0.05522642330710145,
'albumin': 0.05067079561524062,
'protime': 0.0035883401574868462},
'sgot': {'age': 0.0007974932531030433,
'bilirubin': 0.24038403856821702,
'alk_phosphate': -0.05522642330710145,
'sgot': 1.0,
'albumin': -0.08466642672657282,
'protime': -0.02188220220971739},
'albumin': {'age': -0.0821222593960426,
'bilirubin': -0.17056491214839042,
'alk_phosphate': 0.05067079561524062,
'sgot': -0.08466642672657282,
'albumin': 1.0,
'protime': 0.3900284457726809},
'protime': {'age': -0.2135133507321673,
'bilirubin': -0.2069968293170332,
'alk_phosphate': 0.0035883401574868462,
'sgot': -0.02188220220971739,
'albumin': 0.3900284457726809,
'protime': 1.0}}},
'rule_violations': {'outcome': {'violations': 28,
'total': 155,
'violation_rate': 0.18064516129032257},
'sex': {'violations': 15,
'total': 155,
'violation_rate': 0.0967741935483871},
'antivirals': {'violations': 33,
'total': 155,
'violation_rate': 0.2129032258064516},
'malaise': {'violations': 16,
'total': 155,
'violation_rate': 0.1032258064516129},
'anorexia': {'violations': 8,
'total': 155,
'violation_rate': 0.05161290322580645},
'liver_big': {'violations': 5,
'total': 155,
'violation_rate': 0.03225806451612903},
'spleen_palpable': {'violations': 11,
'total': 155,
'violation_rate': 0.07096774193548387},
'ascites': {'violations': 3,
'total': 155,
'violation_rate': 0.01935483870967742},
'varices': {'violations': 4,
'total': 155,
'violation_rate': 0.025806451612903226},
'protime': {'violations': 6,
'total': 155,
'violation_rate': 0.03870967741935484}},
'missingness_fidelity': {'outcome': {'original_missing_rate': 0.0,
'synthetic_missing_rate': 0.0,
'absolute_difference': 0.0},
'age': {'original_missing_rate': 0.0,
'synthetic_missing_rate': 0.0,
'absolute_difference': 0.0},
'sex': {'original_missing_rate': 0.0,
'synthetic_missing_rate': 0.0,
'absolute_difference': 0.0},
'steroid': {'original_missing_rate': 0.0064516129032258064,
'synthetic_missing_rate': 0.0,
'absolute_difference': 0.0064516129032258064},
'antivirals': {'original_missing_rate': 0.0,
'synthetic_missing_rate': 0.0,
'absolute_difference': 0.0},
'fatigue': {'original_missing_rate': 0.0064516129032258064,
'synthetic_missing_rate': 0.0,
'absolute_difference': 0.0064516129032258064},
'malaise': {'original_missing_rate': 0.0064516129032258064,
'synthetic_missing_rate': 0.0,
'absolute_difference': 0.0064516129032258064},
'anorexia': {'original_missing_rate': 0.0064516129032258064,
'synthetic_missing_rate': 0.0,
'absolute_difference': 0.0064516129032258064},
'liver_big': {'original_missing_rate': 0.06451612903225806,
'synthetic_missing_rate': 0.0,
'absolute_difference': 0.06451612903225806},
'liver_firm': {'original_missing_rate': 0.07096774193548387,
'synthetic_missing_rate': 0.01935483870967742,
'absolute_difference': 0.05161290322580645},
'spleen_palpable': {'original_missing_rate': 0.03225806451612903,
'synthetic_missing_rate': 0.0,
'absolute_difference': 0.03225806451612903},
'spiders': {'original_missing_rate': 0.03225806451612903,
'synthetic_missing_rate': 0.0,
'absolute_difference': 0.03225806451612903},
'ascites': {'original_missing_rate': 0.03225806451612903,
'synthetic_missing_rate': 0.0,
'absolute_difference': 0.03225806451612903},
'varices': {'original_missing_rate': 0.03225806451612903,
'synthetic_missing_rate': 0.0,
'absolute_difference': 0.03225806451612903},
'bilirubin': {'original_missing_rate': 0.03870967741935484,
'synthetic_missing_rate': 0.08387096774193549,
'absolute_difference': 0.04516129032258065},
'alk_phosphate': {'original_missing_rate': 0.1870967741935484,
'synthetic_missing_rate': 0.03225806451612903,
'absolute_difference': 0.15483870967741936},
'sgot': {'original_missing_rate': 0.025806451612903226,
'synthetic_missing_rate': 0.0064516129032258064,
'absolute_difference': 0.01935483870967742},
'albumin': {'original_missing_rate': 0.1032258064516129,
'synthetic_missing_rate': 0.07741935483870968,
'absolute_difference': 0.025806451612903222},
'protime': {'original_missing_rate': 0.432258064516129,
'synthetic_missing_rate': 0.3741935483870968,
'absolute_difference': 0.05806451612903224},
'histology': {'original_missing_rate': 0.0,
'synthetic_missing_rate': 0.0,
'absolute_difference': 0.0}},
'privacy_assessment': {'exact_duplicates': 0, 'exact_duplicate_rate': 0.0},
'mixed_type_quality': {'categorical_numeric_preservation': 0.8820386443532539,
'integer_constraint_quality': 0.8788659793814433},
'overall_quality_score': 0.9197601510754626}
Advanced Usage: Customizing Generation Parameters
The framework offers several customization options for different use cases:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
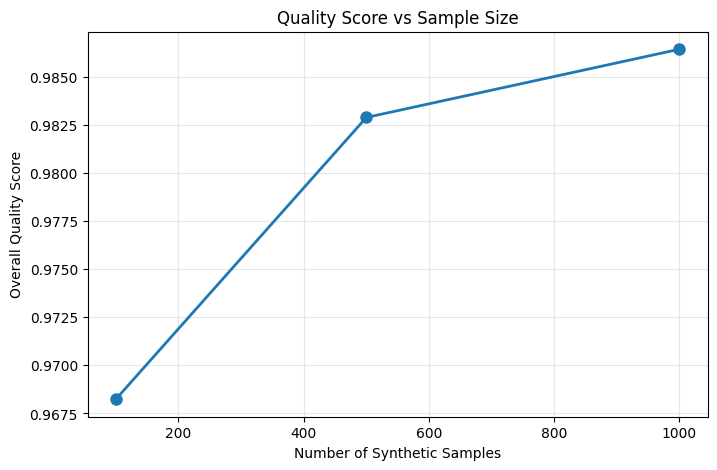
# Example: Generate multiple synthetic datasets with different sizes
sample_sizes = [100, 500, 1000]
quality_scores = []
for size in sample_sizes:
# Generate synthetic data
synth = generator.generate(size)
# Evaluate quality
generator.synthetic_data = synth # Update for evaluation
eval_result = generator.evaluate()
quality_scores.append(eval_result['overall_quality_score'])
print(f"Sample size: {size}, Quality score: {eval_result['overall_quality_score']:.3f}")
# Plot quality vs sample size
plt.figure(figsize=(8, 5))
plt.plot(sample_sizes, quality_scores, 'o-', linewidth=2, markersize=8)
plt.xlabel('Number of Synthetic Samples')
plt.ylabel('Overall Quality Score')
plt.title('Quality Score vs Sample Size')
plt.grid(True, alpha=0.3)
plt.show()
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
Generating 100 synthetic samples...
Generating data using sequential modeling approach...
Generating column: age
Generating column: income
Generating column: education
Generating column: credit_score
Generating column: region
Generating column: account_balance
Generating column: customer_type
Post-processing: Enforcing data types and rules...
Step 1: Enforcing data types...
Step 2: Enforcing deterministic rules...
Applied rule for customer_type
Step 3: Applying missingness patterns...
Applied MAR pattern for education (12.0% missing)
Applied MAR pattern for account_balance (11.0% missing)
Generation completed successfully!
Pillar 5: Evaluating synthetic data quality...
Evaluating statistical fidelity...
Evaluating correlation preservation...
Verifying deterministic rules...
Evaluating missingness fidelity...
Assessing privacy...
Overall Quality Score: 0.968
Sample size: 100, Quality score: 0.968
Generating 500 synthetic samples...
Generating data using sequential modeling approach...
Generating column: age
Generating column: income
Generating column: education
Generating column: credit_score
Generating column: region
Generating column: account_balance
Generating column: customer_type
Post-processing: Enforcing data types and rules...
Step 1: Enforcing data types...
Step 2: Enforcing deterministic rules...
Applied rule for customer_type
Step 3: Applying missingness patterns...
Applied MAR pattern for education (9.0% missing)
Applied MAR pattern for account_balance (7.8% missing)
Generation completed successfully!
Pillar 5: Evaluating synthetic data quality...
Evaluating statistical fidelity...
Evaluating correlation preservation...
Verifying deterministic rules...
Evaluating missingness fidelity...
Assessing privacy...
Overall Quality Score: 0.983
Sample size: 500, Quality score: 0.983
Generating 1000 synthetic samples...
Generating data using sequential modeling approach...
Generating column: age
Generating column: income
Generating column: education
Generating column: credit_score
Generating column: region
Generating column: account_balance
Generating column: customer_type
Post-processing: Enforcing data types and rules...
Step 1: Enforcing data types...
Step 2: Enforcing deterministic rules...
Applied rule for customer_type
Step 3: Applying missingness patterns...
Applied MAR pattern for education (10.1% missing)
Applied MAR pattern for account_balance (8.4% missing)
Generation completed successfully!
Pillar 5: Evaluating synthetic data quality...
Evaluating statistical fidelity...
Evaluating correlation preservation...
Verifying deterministic rules...
Evaluating missingness fidelity...
Assessing privacy...
Overall Quality Score: 0.986
Sample size: 1000, Quality score: 0.986
Saving and Loading Trained Generators
You can save trained generators for later use:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
import pickle
# Save the trained generator
with open("trained_generator.pkl", "wb") as f:
pickle.dump(generator, f)
print("Generator saved to 'trained_generator.pkl'")
# Load and use the saved generator
with open("trained_generator.pkl", "rb") as f:
loaded_generator = pickle.load(f)
# Generate new data with loaded generator
new_synthetic = loaded_generator.generate(100)
print(f"\nGenerated {len(new_synthetic)} new samples with loaded generator")
print("First 3 rows:")
new_synthetic.head(3)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
Generator saved to 'trained_generator.pkl'
Generating 100 synthetic samples...
Generating data using sequential modeling approach...
Generating column: age
Generating column: income
Generating column: education
Generating column: credit_score
Generating column: region
Generating column: account_balance
Generating column: customer_type
Post-processing: Enforcing data types and rules...
Step 1: Enforcing data types...
Step 2: Enforcing deterministic rules...
Applied rule for customer_type
Step 3: Applying missingness patterns...
Applied MAR pattern for education (11.0% missing)
Applied MAR pattern for account_balance (12.0% missing)
Generation completed successfully!
Generated 100 new samples with loaded generator
First 3 rows:
| age | income | education | credit_score | region | account_balance | customer_type | |
|---|---|---|---|---|---|---|---|
| 0 | 34 | 55263 | PhD | 422 | West | 9342.41 | Premium |
| 1 | 53 | 70729 | Bachelor | 531 | North | 4868.31 | Premium |
| 2 | 35 | 54367 | Master | 411 | Central | 420.55 | Premium |
Best Practices and Tips
1. Data Preparation
- Ensure your data has meaningful column names
- Handle extreme outliers before training
- Consider the trade-off between data size and quality
2. Parameter Tuning
- cart_max_depth: Increase for more complex relationships (default: 5)
- rule_threshold: Lower for more flexible rules (default: 0.99)
3. Quality Assessment
- Always evaluate generated data before use
- Check for preservation of key relationships
- Verify privacy protection (no exact duplicates)
4. Use Cases
- Testing and Development: Generate test data that mirrors production
- Privacy Compliance: Share synthetic data instead of real data
- Data Augmentation: Increase dataset size for machine learning
- Education: Provide realistic datasets for teaching
Performance Considerations
Let’s analyze the framework’s performance characteristics:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
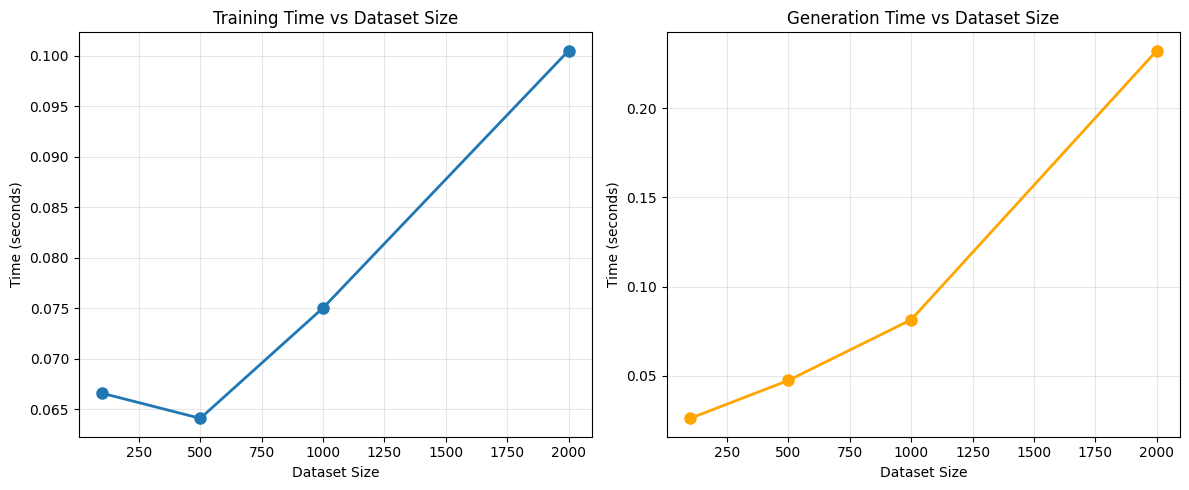
import time
# Test performance with different data sizes
data_sizes = [100, 500, 1000, 2000]
training_times = []
generation_times = []
for size in data_sizes:
# Create data
test_data = create_sample_customer_data(size)
# Time training
gen = SyntheticDataGenerator(verbose=False)
start_time = time.time()
gen.fit(test_data)
training_time = time.time() - start_time
training_times.append(training_time)
# Time generation
start_time = time.time()
_ = gen.generate(size)
generation_time = time.time() - start_time
generation_times.append(generation_time)
print(f"Size: {size:4d} | Training: {training_time:6.3f}s | Generation: {generation_time:6.3f}s")
# Plot performance
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 5))
ax1.plot(data_sizes, training_times, 'o-', label='Training Time', linewidth=2, markersize=8)
ax1.set_xlabel('Dataset Size')
ax1.set_ylabel('Time (seconds)')
ax1.set_title('Training Time vs Dataset Size')
ax1.grid(True, alpha=0.3)
ax2.plot(data_sizes, generation_times, 'o-', label='Generation Time', linewidth=2, markersize=8, color='orange')
ax2.set_xlabel('Dataset Size')
ax2.set_ylabel('Time (seconds)')
ax2.set_title('Generation Time vs Dataset Size')
ax2.grid(True, alpha=0.3)
plt.tight_layout()
plt.show()
1
2
3
4
Size: 100 | Training: 0.067s | Generation: 0.026s
Size: 500 | Training: 0.064s | Generation: 0.047s
Size: 1000 | Training: 0.075s | Generation: 0.081s
Size: 2000 | Training: 0.101s | Generation: 0.232s
Conclusion
This High-Fidelity Synthetic Data Generation Framework provides a sophisticated solution for creating privacy-preserving synthetic datasets. Key advantages include:
- Automatic Discovery: The framework automatically identifies column types, relationships, and patterns
- Preservation of Complex Relationships: Deterministic rules and conditional dependencies are maintained
- Realistic Missing Data: Missing patterns (MAR/MCAR) are learned and replicated
- Privacy Protection: No exact duplicates ensures individual privacy
- Comprehensive Evaluation: Built-in quality assessment with visual reports
Whether you’re working with customer data, clinical records, or any other sensitive information, this framework enables you to generate high-quality synthetic data that maintains the statistical properties of your original dataset while protecting individual privacy.
Next Steps
- Try it with your own data: The framework is designed to work with any tabular dataset
- Customize parameters: Experiment with different CART depths and rule thresholds
- Contribute: The framework is open-source - contributions and feedback are welcome!
For more information, visit the GitHub repository or contact the author, Deniz Akdemir, PhD, at deniz.akdemir.work@gmail.com.
References
- Breiman, L. (2001). “Random Forests.” Machine Learning, 45(1), 5-32.
- Little, R. J., & Rubin, D. B. (2019). Statistical analysis with missing data (Vol. 793). John Wiley & Sons.
- Patki, N., Wedge, R., & Veeramachaneni, K. (2016). “The synthetic data vault.” In 2016 IEEE International Conference on Data Science and Advanced Analytics (DSAA) (pp. 399-410).
- Xu, L., Skoularidou, M., Cuesta-Infante, A., & Veeramachaneni, K. (2019). “Modeling tabular data using conditional GAN.” In Advances in Neural Information Processing Systems (pp. 7335-7345).
- Akdemir, D. (2025). “High-Fidelity Synthetic Data Generation Framework.” GitHub repository.
Blog post created: July 31, 2025